Induction of Recombinant Uridine Phosphorylase and Its Application in Biosynthesis of Pyrimidine Nucleosides*
DING Qingbao (丁庆豹), OU Ling (欧伶), WEI Dongzhi (魏东芝),**, WEI Xiaokun (魏晓琨), XU Yanmei (许彦梅) and ZHANG Chunyan (张春艳)
Induction of Recombinant Uridine Phosphorylase and Its Application in Biosynthesis of Pyrimidine Nucleosides*
DING Qingbao (丁庆豹)1, OU Ling (欧伶)1, WEI Dongzhi (魏东芝)1,**, WEI Xiaokun (魏晓琨)2, XU Yanmei (许彦梅)2and ZHANG Chunyan (张春艳)2
1State Key Lab of Bioreactor Engineering, East China University of Science and Technology, Shanghai 200237, China2Shanghai Pucheng Bioscience and Biotechnology Co., Ltd., Shanghai 200235, China
RecombinantpUDP, which overexpressed uridine phosphorylase (UPase), was constructed. 0.5 mmol·L-1lactose had a similar induction effect as the commonly used inducer IPTG during 2.5-5.5 h of cell growth. The lactose-induced UPase was stable at 50 °C. Wet cells of pUDP was used as catalyst to biosynthesize 5-fluorouridine from 30 mmol·L-1uridine and 5-fluorouracil in phosphate buffer (pH 7.0) catalyzed at 50 °C for 1.5 h and the yield of 5-fluorouridine was higher than 68%. Under the optimum reaction conditions for production of 5-fluorouridine, 5-methyluridine and azauridine were synthesized from uridine by pUDP, the yield was 61.7% and 47.2% respectively. Deoxynucleosides were also synthesized by pUDP, but the yield was only about 20%.
5-fluorouridine, lactose, uridine phosphorylase, pyrimidine nucleoside, uridine, 5-fluorouracil
1 Introduction
Nucleosides and their derivatives are important sources of antiviral and anticancer drugs [1, 2], and many nucleoside drugs, such as zidovudine, ribavirin, lamivudine, and capecitabine, are widely used in clinical practice. Most non-natural nucleosides are usually synthesized chemically, resulting in serious environmental pollution because of the large amounts of toxic and hazardous reagents used.
There are extensive reports on the enzymatic synthesis of nucleosides using nucleoside phosphorylases (NPase) [3]. Many nucleosides have been successfully synthesized enzymatically, such as ribavirin [4, 5], 5-methyluridine [6, 7], adenine arabinoside [8, 9], and dideoxyinosine [10]. The major disadvantage of this process, which limits its application, is the large amount of biomass needed for synthesis.
As one of the NPases, uridine phosphorylase (UPase, EC 2.4.2.4) is an enzyme that participates in the pyrimidine salvage pathway of eukaryotes and prokaryotes. UPase catalyzes the reversible phosphorolysis of pyrimidine nucleosides to the free pyrimidine base and ribose-1-phosphate (R-1-Pi). It has been found that UPase from(.) is a homohexamer, with each subunit being 27150. The use of UPase in the synthesis of nucleoside analogues has been elucidated in scientific publications and patents.
In molecular biology, IPTG is the common inducer for UPase expression by recombinant bacteria. IPTG produces excellent inducement that is sustained for a long period. However, it is too expensive and potentially toxic. Thus, it is not advocated by most of pharmacopoeia. Lactose is a natural substrate of natural enzymes of bacteria controlled byoperon. It is also a native inducer ofoperon. Due to its low cost, harmlessness, and avirulence, lactose is suitable for use as an inducer in most cases [11, 12].
In this study, the.gene encoding UPase is cloned and expressed to construct recombinant.pUDP. Intact recombinant cells are induced with lactoseand used as UPase to biosynthesize 5-fluorouridine and other nucleosides.
2 Materials and methods
2.1 Materials
Nucleosides and bases, such as thymine, uracil, cytosine, 5-fluorouracil, azauracil, 5-fluorocytosine, thymidine, 2′-deoxyuridine, cytidine, 5-methyluridine and uridine, were supplied by Shanghai Biocaxis Chemicals Co., Ltd. IPTG was purchased from Sigma, USA, tryptone and yeast extract were from Oxoid, UK, and low molecular protein marker was from Tiangen Biotechnology Co. Other reagents and solvents were of commercial quality and were used without further purification. Cloning vector pMD-18T and plasmid expression vector pET-11a were from Novagen. Strain.JM109 and.BL21 (DE3) were stored in the laboratory.DNA polymerase, T4 ligase, and all restriction enzymes used in DNA cloning were purchased from Takara (Japan), while the GeneClean kit for DNA purification was purchased from Generay Biotech (Shanghai).
2.2 Methods
2.2.1
All DNA manipulations were performed using standard procedures [13]. The gene encoding UPase, referred to as, was amplified from.genome DNA by polymerase chain reaction (PCR) with the synthetic primers (+)5′-GGGAATTCCATATGTCCAAGTCTGATG and (-)5′-GCGGATCCTTACAGCAGACGACGCGCC. Amplification was performed usingpolymerase with 30 cycles, each at 94 °C for 30 s, 55 °C for 60 s, and 72 °C for 1 min. PCR products were detected and separated with 0.9% agarose gels. The target strand was recovered and purified, then inserted into pMD18-T vector. Nucleotide sequence was determined with Invitrogen (Shanghai). Assembly and analysis of DNA sequences were carried out by Lasergene (Version 7.1.0, DNASTAR, Inc.). The BLAST Web site was used for database search. Once thegene was identified, it was digested withI andI restriction endonucleases to be isolated from pMD18-T vector, then cloned to expression vector pET-11a to form recombinant plasmids pET-11a-. pET-11a-was transformed into.BL21 (DE3) to construct recombinant strain pUDP, which can be expressed to obtain large amounts of UPase.
2.2.2
The Luria-Bertani (LB) culture medium contained 10 g peptone, 5 g yeast extract, 5 g NaCl, and distilled water to form a total volume of 1 L. It was adjusted to pH 7.0 with 2 mol·L-1NaOH. 30 ml medium was inoculated into 250 ml flasks with one loopful of cells of pUDP subcultured on agar plates of the same medium with 50 μg·ml-1sodium carbenicillin. After aerobic cultivation at 37 °C for 15 h, 1.0 ml of the cultured broth was transferred to 30 ml of the same medium in 250 ml flasks, followed by aerobic cultivation at 37 °C for 3-4 h. When OD660reached 0.6 (usually about 3-4 h), 0.5 mmol·L-1lactose or IPTG was added into the broth, and the strains were cultured at 37 °C for another 6 h. The cells were then harvested by centrifugation (10000, 15 min) and washed twice with 100 mmol·L-1Tris-HCl buffer with pH 7.0. The cells were stored at-20 °C and used as intact cells for reactions.
2.2.3
UPase was assayed according to the method described by Saunders[14]. Briefly, an assay solution containing 2 mg wet cell paste, 10 mmol·L-1uridine, and 100 mmol·L-1potassium phosphate buffer (pH 7.0) in a total volume of 2 ml was incubated at 60 °C for 20 min. The reaction was terminated by adding 1 mol·L-1ice-cold NaOH (2 ml). The reaction mixture was centrifuged and the supernatant was diluted 50-fold with NaOH (pH 12).
Enzymatic activity was determined spectrophotometrically by measuring the absorption at 290 nm relative to a blank. The blank solution was similar to the assay solution except that the cells were added after the addition of NaOH. Under the above reaction conditions, the specific activities of enzymes were expressed as a 0.01 change in absorption at 290 nm of the reaction mixture per gram wet cell paste.
2.2.4
SDS-PAGE analysis was performed as described in Ref. [13].
2.2.5
The standard enzymatic reaction liquid (5 ml) contained 30 mmol·L-1nucleoside and base, 50 mmol·L-1potassium phosphate buffer with pH 7.0, and 15 mg wet cells. It was shaken at 50°C for 1.5 h. Once the reaction was complete, the liquid was heated in a 100 °C water bath for 5 min, and the cells were centrifuged at 4000 r·min-1for disposal. The supernatant liquid was diluted 1000-fold with water and products were detected with HPLC.
2.2.6
All products were analyzed at room temperature (25 °C) by high performance liquid chromatography (HPLC Angilent 1200) with UV detection at 254 nm using a Hypersil ODS2 5 µm column (4.6 mm×250 mm) with a solvent of 5% (by volume) acetonitrile and 95% (by volume) water at a flow rate of 1.0 ml·min-1.
3 Results and discussion
3.1 UPase overexpression by recombinant pUDP
To obtain strains containing a high activity of UPase, the expression system including the plasmid of pET-11a and host strain.BL21 (DE3), which is regulated by IPTG induced-transcription, was used. UPase was significantly overexpressed by recombinant pUDP after IPTG inducement (Fig. 1). No inclusion bodies were found in pUDP and most expressed UPase was soluble.

Figure 1 SDS-PAGE result of pUDP expression product
M—protein marker; 1—supernate after the total cell centrifugation; 2—Precipitate after sonication; 3—supernate after sonication; 4—control of.BL21 (pET-11a) after induction; 5,6,7,8—induction for 4, 3, 2, and 1 h, respectively; 9—sample before adding IPTG
3.2 Cell growth with lactose and IPTG
After cell culture for 3 h, with OD660about 0.6, 0.5 mmol·L-1lactose or IPTG was added into the broth. The growth curve of recombinant pUDP was determined and compared with the control (no inducer). The results are shown in Fig. 2. The bacterium growth rate drops slightly once IPTG is added into the broth, whereas the use of lactose as inducer does not significantly hamper the cell growth, with growth rates approximately similar to those of the control. As a strong inducer, IPTG enters cells and commands their protein expression system to express foreign enzyme protein, so that the majority of the nutrients in the medium are used for protein expression, thus inhibiting the cell growth.
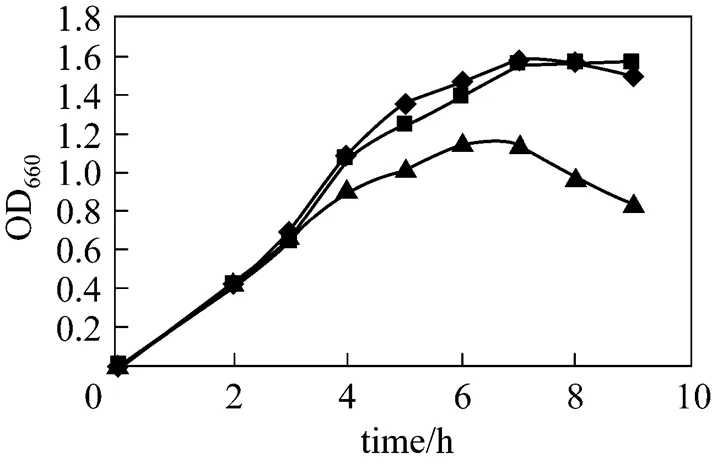
Figure 2 Influence of inducers on the growth of strains
◆ control;■ lactose;▲ IPTG
3.3 Activity of UPase induced at different time by the inducer
A total concentration of 0.5 mmol·L-1lactose or IPTG was added into the broth in different periods of bacterial growth. All inductions were carried out at 37 °C for 6 h before determining the activity of UPase. Table 1 presents the results. The highest activity of UPase was obtained when the cells were cultured for 3.5 h. At this time, the cells were in prophase and the growth was logarithmic because of the state of division and enough nutrition in the medium, so that the addition of inducer resulted in optimal expression protein. With IPTG, the initial induction time was relatively short, only 1 h. After this period, IPTG induction decreased considerably. With lactose, the induction time was a little longer than that of IPTG.. Lactose exhibited good induction, with a high growth rate between 3.5-5.5 h. If pUDP was induced after 5.5 h culture, lactose also had a little induction, but UPase activity was very low. This is because the nutrient in the medium had already been consumed by the recombinant, and little was left to support protein expression.
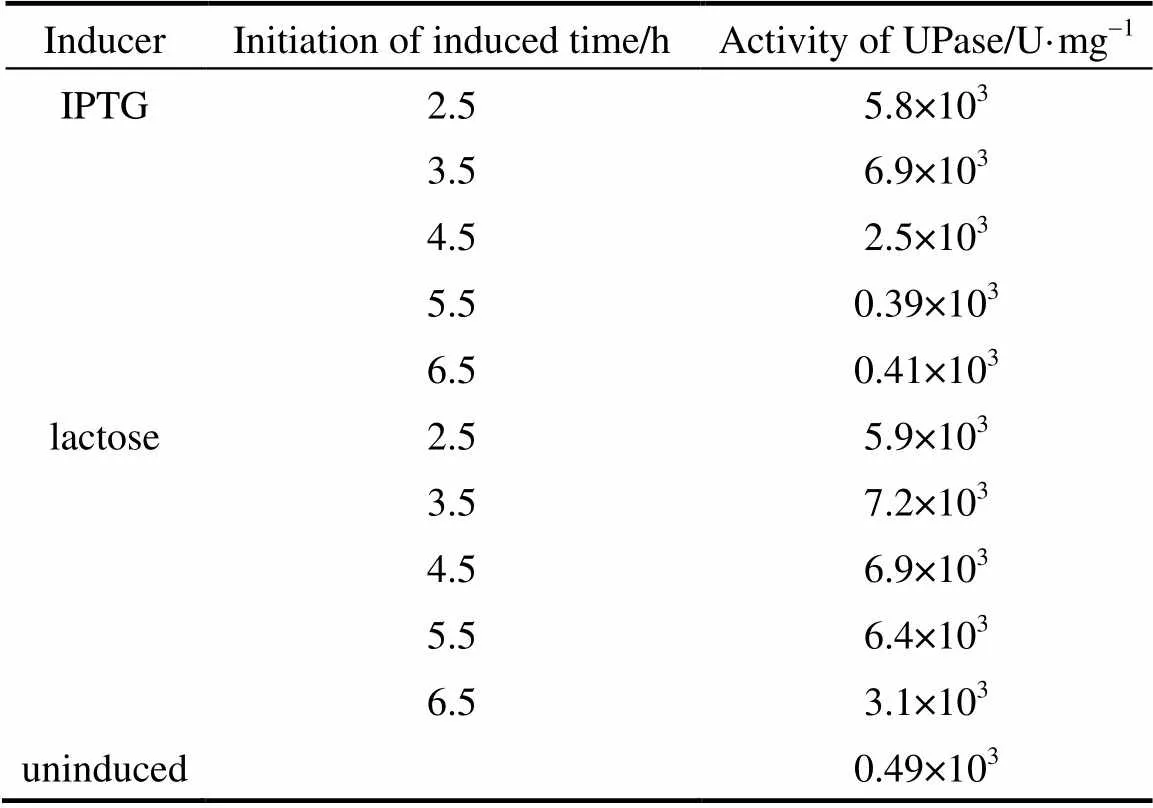
Table 1 Activity of UPase induced at different time byIPTG and lactose
3.4 Activity of UPase at different concentrations of lactose
Different concentrations of lactose were used to induce pUDP and 0.5 mmol·L-1IPTG was used as control. The results of activity and SDS-PAGE are shown in Table 2 and Fig. 3, respectively. UPase activity assay shows that the increase in concentration of lactose up to 2 mmol·L-1increases the biomass. At higher lactose concentration, the enzyme activity and biomass do not increase significantly. Very low concentrations of lactose have good inducement. The results of SDS-PAGE are consistent with UPase activity.
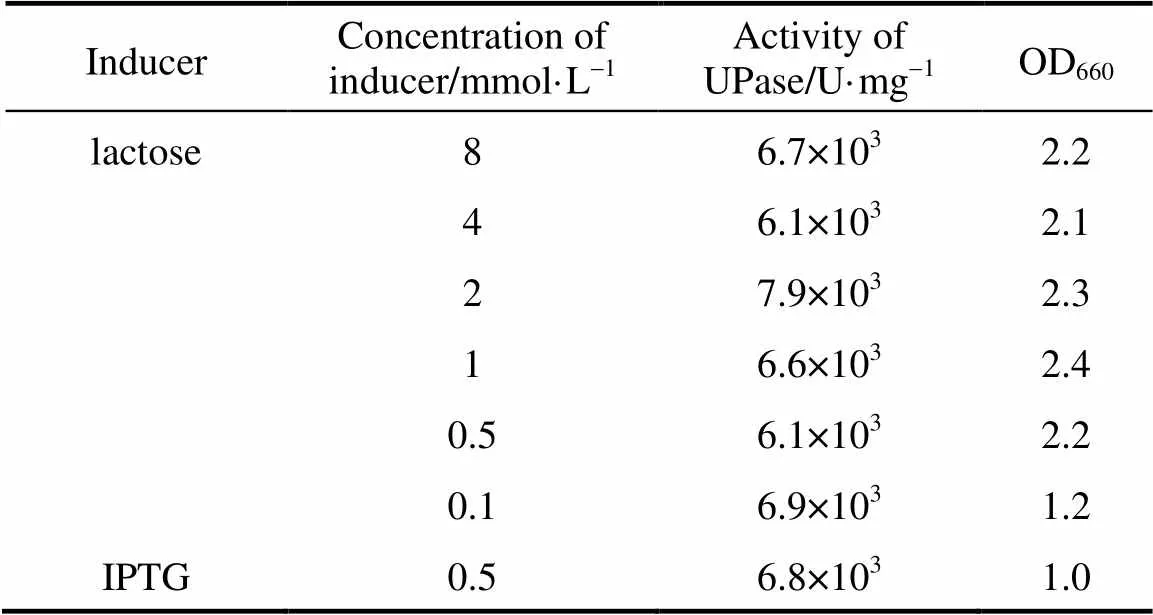
Table 2 UPase induced by differentconcentrations of lactose
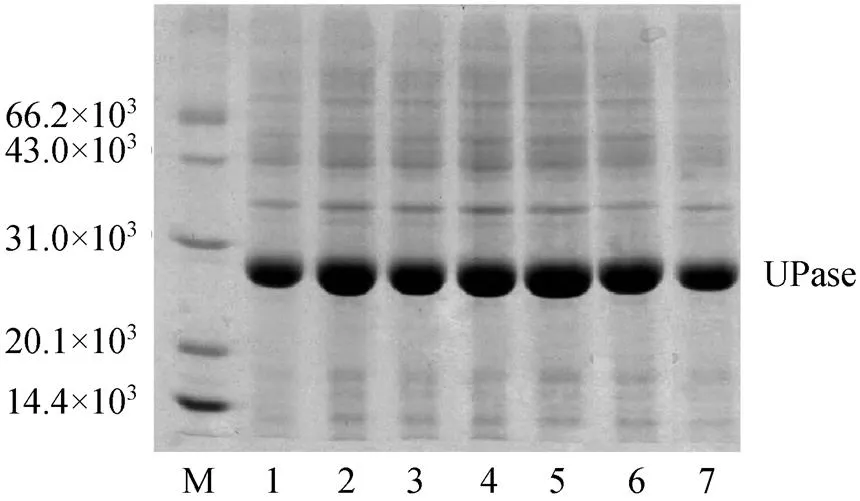
Figure 3 SGS-PAGE of UPase induced by different concentrations of lactose
M—protein marker; lanes 1—6: lactose concentration 8, 4, 2, 1, 0.5, and 0.1 mmol·L-1, respectively; lane 7: UPase induced by IPTG
3.5 Effect of induction time on the activity of UPase
The activity of UPase was measured after it was induced with lactose for different durations (4, 5, 6, 7, and 8 h). The results are listed in Table 3. As the induction time is longer, the activity of UPase increases gradually, reaching a peak at 6 h. When the induction time is too short, the target protein is not fully expressed. When the induction time is longer than 6 h, the UPase activity declines a little, because bacterial growth is in the wane phase and the concentration of the biomass declines. Thus, the optimum induction time is 6 h, at which the target protein is fully expressed, bacterial growth is stable, and the highest concentration of biomass is achieved.
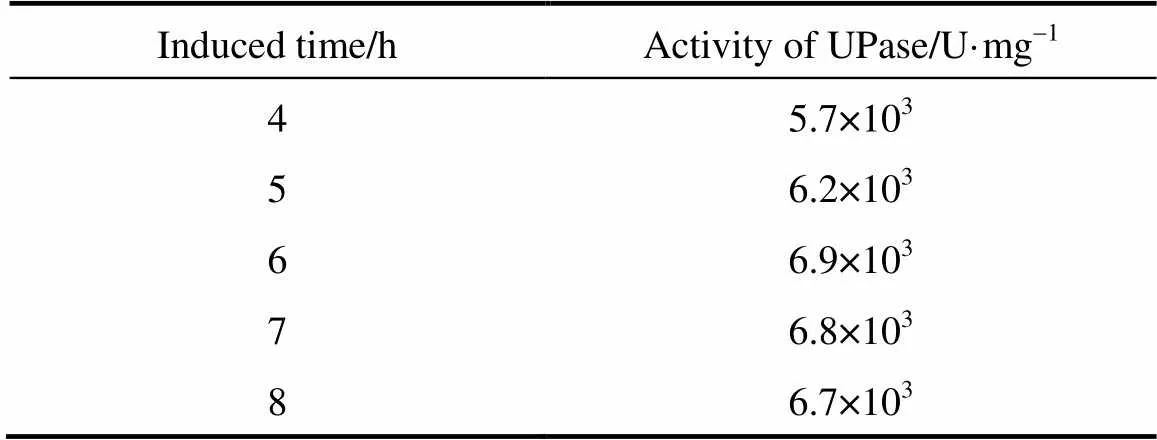
Table 3 Activity of TPase induced for different durations
3.6 Thermal stability of UPase
After lactose induction, the wet cells of pUDP were heated to 50, 60, 65, and 70 °C for 10, 20, 30, 40, 60, and 120 min before determination of UPase activity. Fig. 4 presents the results. UPase is very stable at 50 °C and a little unstable at 60 °C compared with the result at two other temperatures. At 70 °C, the activity of UPase decreases quickly. Thus, 50 °C is selected for the succeeding nucleoside biosynthesis.
3.7 Biosynthesis of 5-fluorouridine with UPase
5-fluorouridine (5-FUR), a key intermediate of the anticancer drug Furtulon, was synthesized with intact wet cells of pUDP as the catalyst. 5-fluorouracil (5-FU) was used as ribosyl acceptor and uridine (UR) was used as ribosyl donor. The reaction mechanism is shown in Fig. 5.
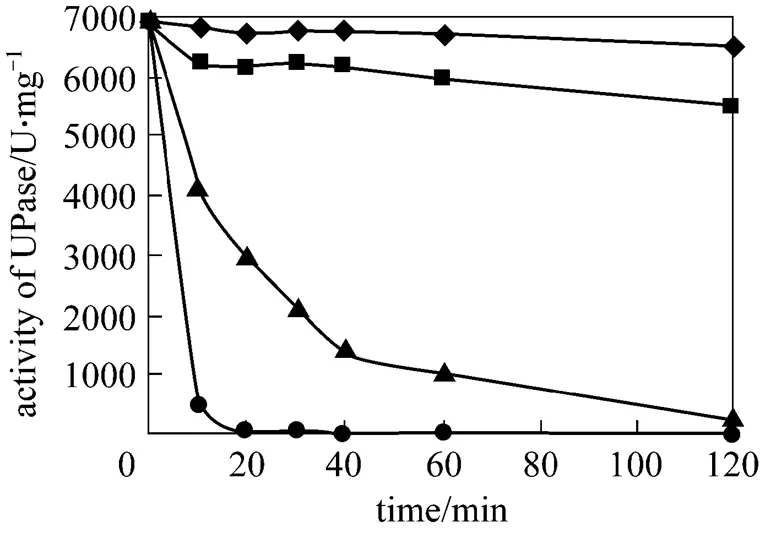
Figure 4 Stability of UPase at different temperatures
◆ 50 °C;■ 60 °C;▲ 65 °C;● 70 °C
The reaction process using the standard reaction is shown in Fig. 6. The yield of 5-FUR reaches 68% in 1-2 h, but it decreases later, mainly due to the formation of intermediate product ribose-1-phosphate, which is less stable, from the decompose of nucleoside in the reversible reaction catalyzed by nucleoside phosphorylase. In order to obtain higher yield, the reaction time should be limited to1-2 h. For convenience, the reaction was terminated at 1.5 h in the following reactions.
Figure 7 shows the effect of temperature on 5-FUR yield measured with HPLC for 1.5 h reaction time when all other conditions are kept constan. The optimum yield is achieved in the range of 45-60 °C.
5-FUR was biosynthesized at different pH according to the standard reaction. The effect of pH on the yield is shown in Fig. 8. The optimum pH is between 7.0-7.5.
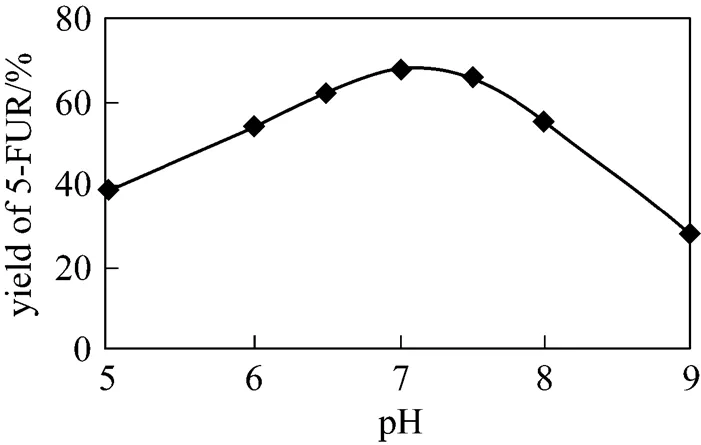
Figure 8 Effect of pH value on the yielde of 5-FUR
Figure 9 Effect of amount of cells on the yield of 5-FUR
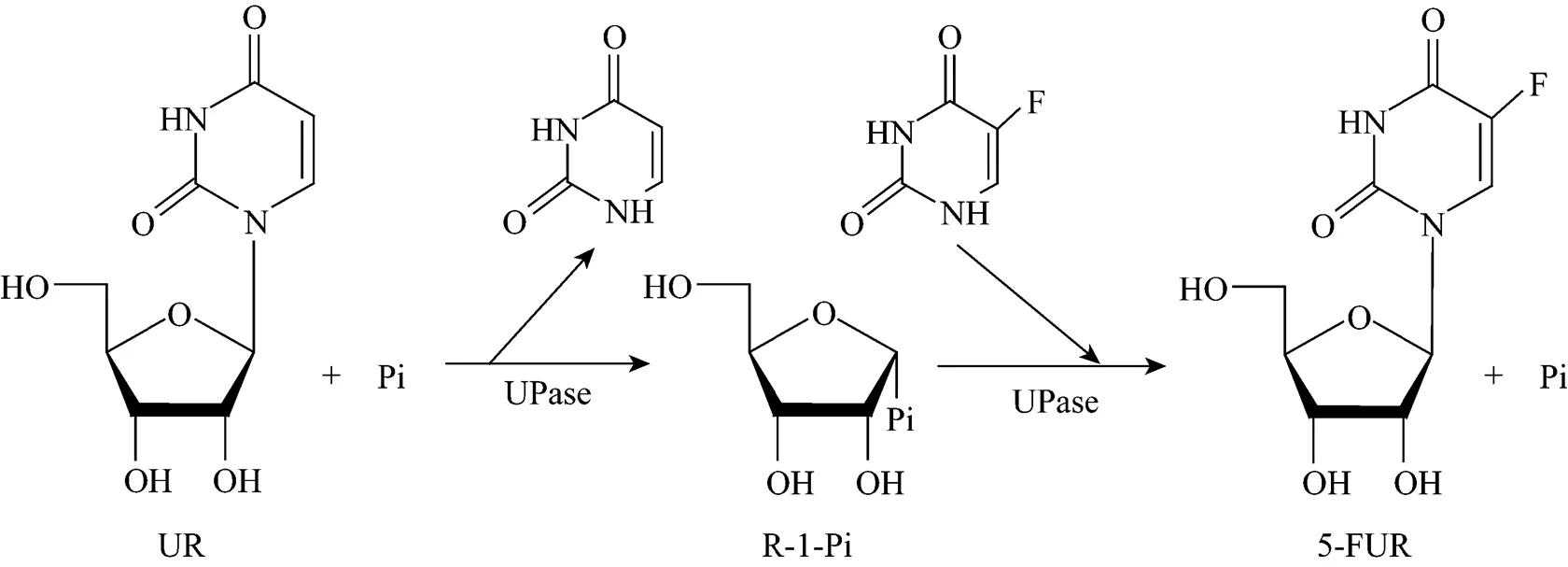
Figure 5 Proposed mechanism for the production of 5-FUR
Figure 6 Effect of reaction time on the yield of 5-FUR
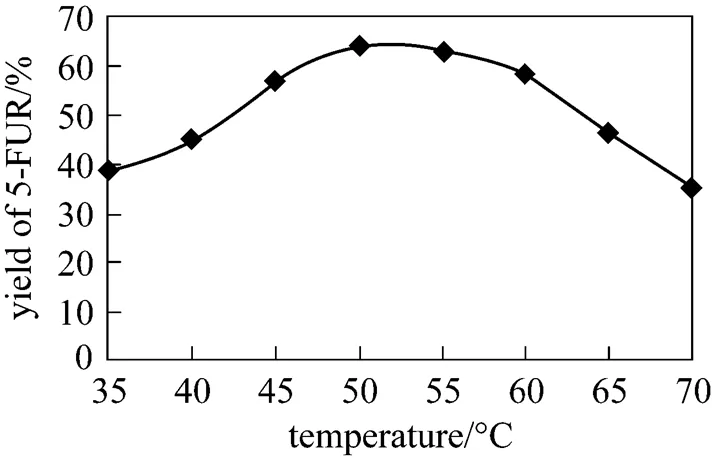
Figure 7 Effect of reaction temperature on the yield of 5-FUR
Under the standard reaction conditions, different amounts of wet intact cells were added as catalyst. After reaction at 50 °C for 1.5 h, the yield of 5-FUR was measured with HPLC. Fig. 9 shows the effect of amount of wet cells on the yield. The optimum amount is between 10-20 mg.
3.8 Biosynthesis of other pyrimidine nucleosides with UPase
According to the above data, the optimum reaction conditions are as follows: 15 mg wet intact cells (pUDP), pH 7.0 phosphate buffer solution, 50 °C, reaction time 1.5 h, and total volume of 5 ml. Under these reaction conditions, different nucleosides and bases listed in Table 4 were used in the reaction system. The nucleosides, except cytidine and its derivative, were transformed by recombinant bacteria with good yield. Cytosine and its analogues are not substrates of UPase, so cytidine and 5-fluorocytidine are not biosynthesized with thymine added [15, 16]. Cytidine was not detected in the reaction solution after reaction was terminated. However, it is possible for a small amount of cytidine to transform to uridine by cytidine deaminase, which is converted to 5-methyluridine, because a large amount of uridine is detected in the reaction mixture. As seen in Table 4, UPase is expressed by pUDP synthesized deoxynucleotides, such as 2′-deoxy-5- fluorouridine from 2′-deoxyuridine or thymidine, but the yield is low.
4 CONCLUSIONS
Thegene from.was cloned and linked to pET-11a plasmid and transformed into.BL21 (DE3) to construct pUDP. The recombinant bacteria can overexpress a large amount of UPase in soluble form after induction by IPTG. Lactose can be used as an inducer to substitute IPTG, an expensive inducer. High activity of UPase can be achieved with 7900 U·mg-1wet cells as catalyst. UPase has good stability at 50 °C.
The reaction conditions of nucleoside biosynthesis by pUDP were demonstrated using synthesis of 5-fluorouridine. The highest yield was obtained with the reaction time of 1-2 h at 45-60 °C and pH 7.0-7.5. The optimum amount of wet cells was between 10-20 mg in 5 ml reaction solution and increase in the amount did not lead to a higher yield.
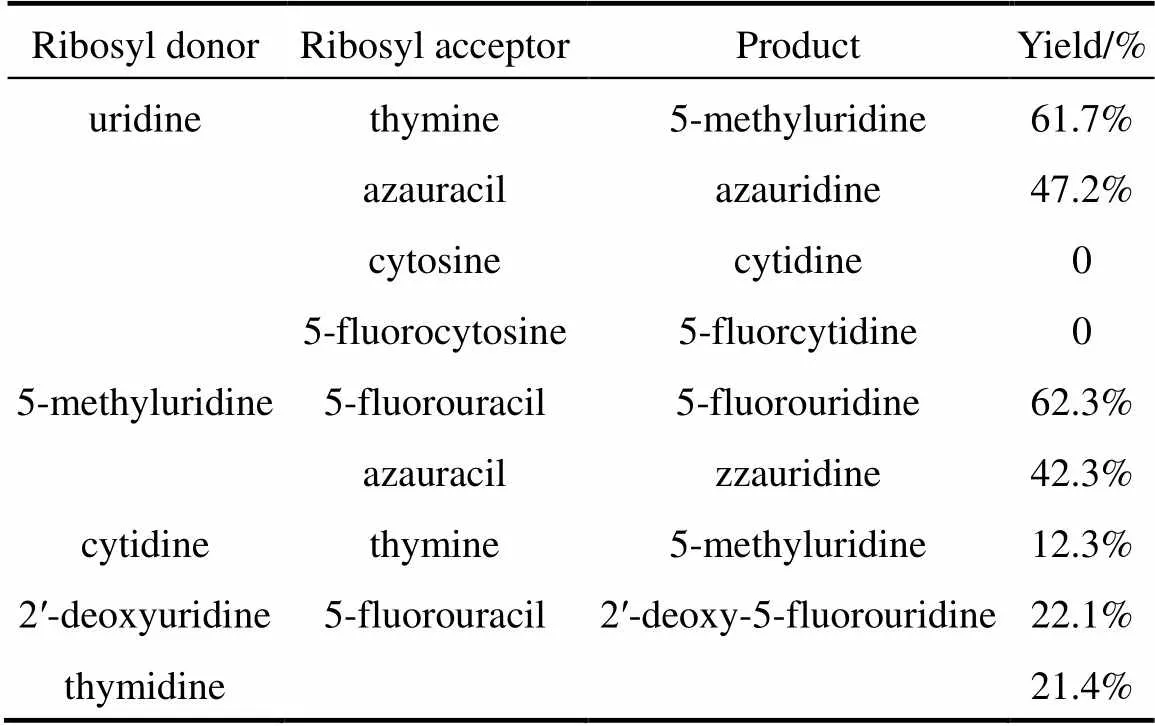
Table 4 Conversion of other pyrimidine nucleosides withrecombinant UPase
Various nucleosides, except cytidine and its analogues, can be biosynthesized by pUDP with good yields. Since cytosine is not a substrate of UPase, cytidine and analogues form. UPase is not appropriate to synthesize deoxynucleotides because the yield is low.
1 Galmarini, C.M., Mackey, J.R., Dumontet, C., “Nucleoside analogues and nucleobases in cancer treatment”,, 3, 415-423 (2002).
2 De Clercq, E., “Antiviral drugs: Current state of the art”,..., 22 (1), 73-89 (2001).
3 Hanrahan, J., Hutchinson, D., “The enzymatic synthesis of antiviral agents”,.., 23, 193-210 (1992).
4 Utagawa, T., Morisawa, H., “Enzymatic synthesis of virazole by purine nucleoside phosphorylase of enterobacterium aerogens”,..., 50, 121-126 (1986).
5 Shirae, H., Yokozeki, K., “Enzymatic production of ribavirin from purine nucleosides byATCC954”,..., 52 (7), 1777-1783 (1988).
6 Hori, N., Watanabe, M., Yamazaki, Y., Mikami, Y., “Synthesis of 5-methyluridine by a thermophile,JTS 859”,..., 53 (1), 197-202 (1989).
7 Ishii, N., Shirae, H., Yokozeki, K., “Enzymatic production of 5-methyluridine from purine nucleosides and thymine byAJ-2992”,..., 52 (12), 3209-3218 (1989).
8 Utagawa, T., Hirokazu, M., Fumihiro, Y., Akihiro, Y., Koji, M., “Microbiological synthesis adenine arabinoside”,..., 49 (4), 1053-1058 (1985).
9 Wei, X.K., Ding, Q.B., Zhang, L., “Induction of nucleoside phosphorylase inand enzymatic synthesis of adenine arabinoside”,..., 9 (7), 520-526 (2008).
10 Shirae, H., Kobayashi, K., Shiragami, H., Irie, Y., Yasuda, N., Yokozeki, K., “Production of 2′,3′-dideoxyadenosine and 2′,3′-dideoxyinosine from 2′,3′-dideoxyuridine and the corresponding purine bases by resting cells ofAJ2595”,..., 55 (2), 410-424(1989).
11 Donovan, R.S., Robinson, C.W., Glick, B.R., “Review: Optimizing inducer and culture conditions for expression of foreign proteins under the control of the lac promoter”,..., 16, 145-154 (1996).
12 Monteiro, R.A., Souza, E.M., Yates, M.G., “Use of lactose to induce expression of soluble NifA protein domainin”,..., 46 (11), 1087-1090 (2000).
13 Sambrook, J., Fritsch, E.F., Maniatis, T., Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, NY(1989).
14 Saunders, P.P., Barbara, A.W., Saunders, G.F., “Purification and comparative properties of a pyrimidine nucleoside phosphorylase from”,..., 244 (13), 3691-3697 (1969).
15 Ling, F., Inoue, Y., Kimura, A., “Purification and characterization of a novel nucleoside phosphorylase from asp. and its use in the enzymatic production of adenine arabinoside”,..., 56 (12), 3830-3834 (1990).
16 Benjamin, C.R., Patricia, A.H., “Purification and properties of purine nucleoside phosphorylase from”,..., 248 (6), 2040-2043 (1973).
** To whom correspondence should be addressed. E-mail: dzhwei@ecust.edu.cn
2010-02-01,
2010-11-27.
“Production, Education & Research” item of Shanghai Baoshan (08-H-4).
 Chinese Journal of Chemical Engineering2011年1期
Chinese Journal of Chemical Engineering2011年1期
- Chinese Journal of Chemical Engineering的其它文章
- Effect of Boundary Layers on Polycrystalline Silicon Chemical Vapor Deposition in a Trichlorosilane and Hydrogen System*
- Experimental and CFD Study on the Role of Fluid Flow Pattern onMembrane Permeate Flux
- Separation of Eu3+ Using a Novel Dispersion Combined LiquidMembrane with P507 in Kerosene as the Carrier*
- Fabrication of SPES/Nano-TiO2 Composite Ultrafiltration Membrane and Its Anti-fouling Mechanism*
- Adsorption and Ozonation Kinetic Model for PhenolicWastewater Treatment*
- Properties of Bio-oil from Fast Pyrolysis of Rice Husk*
