Comparative analysis of nuclear proteomes in mitochondrial DNA-depleted A549 cells and their parental cells
ZHAO Peng,ZHANG Zhao-hui,2,ZHONG Wei-jian,YING Xian-ping,YUAN Zhun,YAO Bi-yun,FU Juan-ling,ZHOU Zong-can
(1.Department of Toxicology,Health Science Center,Peking University,Beijing100191,China;2.Institute of Environmental Medicine and Radiation Hygiene,University of South China,Hengyang421001,China;3.Department of Toxicology,Shanghai Municipal Center for Disease Control&Prevention,Shanghai200336,China)
Comparative analysis of nuclear proteomes in mitochondrial DNA-depleted A549 cells and their parental cells
ZHAO Peng1,ZHANG Zhao-hui1,2,ZHONG Wei-jian3,YING Xian-ping3,YUAN Zhun1,YAO Bi-yun1,FU Juan-ling1,ZHOU Zong-can1
(1.Department of Toxicology,Health Science Center,Peking University,Beijing100191,China;2.Institute of Environmental Medicine and Radiation Hygiene,University of South China,Hengyang421001,China;3.Department of Toxicology,Shanghai Municipal Center for Disease Control&Prevention,Shanghai200336,China)
OBJECTIVETo investigate the nuclear proteomes in mitochondrial DNA(mtDNA)-depleted A549 cells(Rho0cells)and their parental cells(Rho+cells),and to learn more about the nuclear responses to mitochondrial dysfunction.METHODSThe nuclear proteomes of Rho0and Rho+cells were characterized by two dimensional electrophoresis(2-DE)and SELDI-TOF Protein Chip technologies,the differentially expressed proteinspots were identified by MALDI-TOF mass spectrum(MS),the nucleophosmin and P53 expression were detected by Western blotting assay,and the mitochondrial membrane potential(MMP)was measured by the laser scanning confocal microscope.RESULTS2-DE results showed 11 protein-spots were down-regulated and 21 protein-spots were up-regulated in Rho0cell nuclei.SELDI-TOF MS analysis with NP20 ProteinChips revealed 4 protein-peaks decreased in Rho0cell nuclei.One down-regulated protein-spot was identified as eIF-6,and 4 up-regulated proteinspots were identified as nucleophosmin,SFRS1,SFRS3 and hnRNP G,respectively.The increased expression of nucleophosmin in Rho0cells was verified by Western blotting.Based on the clues from proteomic analysis,P53 expression in Rho0cells was higher than in Rho+cells,and MMP was consistent in Rho+and Rho0cells.CONCLUSIONmtDNA-depletion induces nuclear proteome alteration.Rho0cells can be used as a model to study the crosstalk between mitochondrion and nucleus.
nuclear proteomics;genes,p53;membrane potential,mitochondrial
Mitochondria play crucial roles in the physiology of most eukaryotic cells.They not only act as the central energy-generating system,but also participate in intermediary metabolism,calcium signaling,and apoptosis[1].Like the nucleus,mitochondrion maintains its own DNA genome,a 16 kilobase circular mitochondrial DNA(mtDNA)genome which contains 37 genes.The mtDNA genome encodes protein subunits of respiratory complexesⅠ,Ⅲ,Ⅳ,andⅤthat are essential for mitochondrialrespiratory function.Additionally,it encodes 22 mitochondrial tRNAs and 2 rRNAs that are required for mitochondrial protein synthesis[2].The communication between mitochondrion and nucleus is of great significance in cell life.On one hand,a majority of mitochondrial proteins are encoded by the nuclear genome and imported into the mitochondria[2],on the other hand,nuclear gene expression can be influenced by signals coming from mitochondria,known as retrograde communication[3-4].Depending on mitochondrial retrograde communication,cells may reconfigure metabolism to accommodate themselves to defects in mitochondria by changing nuclear gene expression[4].mtDNA-depleted human cells will conduce to studying nuclear responses to mitochondrial dysfunction.It has been reported that the expression of a number of nuclear genes that range from transcription factors to genes involved in metabolism,cell architecture and signaling is altered in response to the absence of mtDNA in breast cancer cells[5].To study the cross-talk between mitochondrion and nucleus,mtDNA-depleted A549 cell line was generated in our laboratory according to the protocol described previously[5].Briefly,A549 cells were treated with ethidium bromide 50 μg·L-1for 2 months before one of the several monoclonal cell lines selected by limiting dilution method was successfully amplified and identified as mtDNA-depleted cell line denoted Rho0cell line in this paper.Previous studies demonstrated that Rho0derivative experience resulted in complete depletion of mtDNA,an abnormal mitochondrial structure and impaired mitochondrial function.In this study,to find out more about the nuclear responses to mitochondrial dysfunction,the nuclear proteomes of mtDNA-depleted A549 cells(Rho0cells)and their parental cells(Rho+cells)were investigated by two dimensional electrophoresis(2-DE),matrix-assisted laser desorptionlionization-time of flight(MALDI-TOF)mass spectrum(MS)and surface enhanced laser desorptionlionization(SELDI)-TOF ProteinChip technologies.
1 MATERIALS AND METHODS
1.1 Cell culture and chemicals
Human lung adenocarcinoma A549 cells denoted,Rho+cells obtained from ATCC,were maintained in monolayer culture at 37℃and 5%CO2in F12-k medium supplemented with 10%heat-inactivated fetal bovine serum,penicillin 10 kU·L-1and streptomycin 10 kU·L-1.Rho0cells were maintained under the same conditions as their parental cells besides being supplemented with uridine 50 mg·L-1.Reagents for cell culture were purchased from Gibco,and other reagents used in this work were from Sigma.
1.2 Nuclei purification and protein extraction
Purification of nuclei was performed as described previously[6],and the purity and integrity of cells and isolated nuclei were determined by May-Grünwald-Giemsa staining assay.The isolated nuclei were solubilized in rehydration/lysis buffer containing urea 7 mol·L-1,thiourea 2 mol·L-1,4%(W/V)CHAPS,DTT 50 mmol·L-1,0.2%(W/V)Bio-Lyte 3/10 ampholytes,and 0.002%(W/V)bromophenol blue.The solution was thoroughly mixed by repeated pepetting and incubated at 4℃for 25 min.After centrifugation at 12 000×g and 4℃for 10 min,the supernatant containing the dissolved protein was ready for 2-DE separation.
1.3 Two-dimensional gel electrophoresis
Linear pH gradient IPG strips(7 cm,pH 4-7)were used in 2-DE,and experiments were carried out as described previously[7].The gels were fixed in 40%(V/V)ethanol and 10%(V/V)acetic acid for 1 h,stained with Sypro Ruby for a minimum of 3 h and washed in 10%(V/V)ethanol and 7%(V/V)acetic acid for 1 h before being visualized on FLA-5100 imaging system.2-DE maps of each sample from 3 independent experiments were analyzed using PDQuest software version 8.0.
1.4 SELDI-TOF Protein Chip analysis
SELDI-TOF Protein Chip analysis was performed as described previously[8].The nuclear extracts(protein 3 μg,2 μl)were loaded onto a spot of a NP20 Protein Chip and dried in the air prior to washing three times in MilliQ water for 2 min each time.The spots were dried in the air again,and then two 1 μl aliquots of saturated sinapinic acid solution in 50%(V/V)acetonitrile and 0.5%(V/V)trifluoroacetic acid were applied to each spot,with air-drying between applications.APBS-Ⅱmass spectrometer was used for recording the time-of-flight spectra at laser intensities of 155,a high relative molecular mass(Mr)of 20 ku,and a detector sensitivity of seven.Each spot was analyzed by averaging 65 laser shots from various regions of the spot surface.The peaks were analyzed by Protein Chip software version 3.0.
1.5 Protein identification
In-gel trypsin digestion of proteins,and MALDI-TOF MS analysis were carried out as described previously[9].The peptide mass fingerprinting(PMF)data were subjected to a Protein Data Bank search(Mascot search engine).Up to one missed tryptic cleavage was considered and a mass accuracy of 50×10-6was used for all the tryptic-mass searches.Calculated pI and molecular mass data were obtained by Mascot.
1.6 Western blotting analysis
Logarithmic growth phase Rho+and Rho0cells were lysed in cell lysis buffer(Tris-HCl 50 mmol·L-1pH 7.4,NaCl 250 mmol·L-1,NaF 50 mmol·L-1,Na3VO41 mmol·L-1,EDTA 5 mmol·L-1,0.1%(V/V)Triton X-100,DTT 1 mmol·L-1,PMSF 1 mmol·L-1,aprotinin 1 mg·L-1).Soluble proteins were recovered by centrifugation at 4℃and 12 000×g for 20 min and assayed for protein concentration using the Bradford method.Cell extracts containing 30 μg proteins were separated on 10%SDS-PAGE gels and then transferred to nitrocellulose membranes that were incubated with nucleophosmin or P53 antibodies(at 1∶200 dilutions)purchased from Santa Cruz Biotechnology overnight and subsequently with horseradish peroxidase conjugated secondary antibody for 1 h.Bands were visualized by Western Blotting Luminol Reagent and exposure to Kodak film.The images were acquired by AGFA DuoScan T1200 scanner and analyzed by Quantity One software.
1.7 Determination of mitochondrial membrane potential(MMP)
Logarithmic growth phase Rho+and Rho0cells attached to coverslips were washed once with pre-warmed phosphate buffered saline(PBS)and subsequently incubated with rhodamine123(10 mg·L-1)and Hoechst 33342(1 μmol·L-1)in PBS for 30 min at 37℃in the dark.After incubation,the cells were rinsed twice with PBS,and then analyzed under Leica TCS SP2 laser scanning confocal microscope.The fluorescence intensity of rhodamine123 in cells represents the mitochondrial membrane potential.
1.8 Statistical analysis
2 RESULTS
2.1 Integrity and purity of isolated nuclei
Determination of the purity and integrity of isolated nuclei was performed by light microscopy using May-Grünwald-Giemsa staining assay.As shown in Fig.1 ,preparations of both Rho+and Rho0cell nuclei appeared intact and pure without visible cytoplasm contamination.Each nuclear preparation was verified using this staining assay.
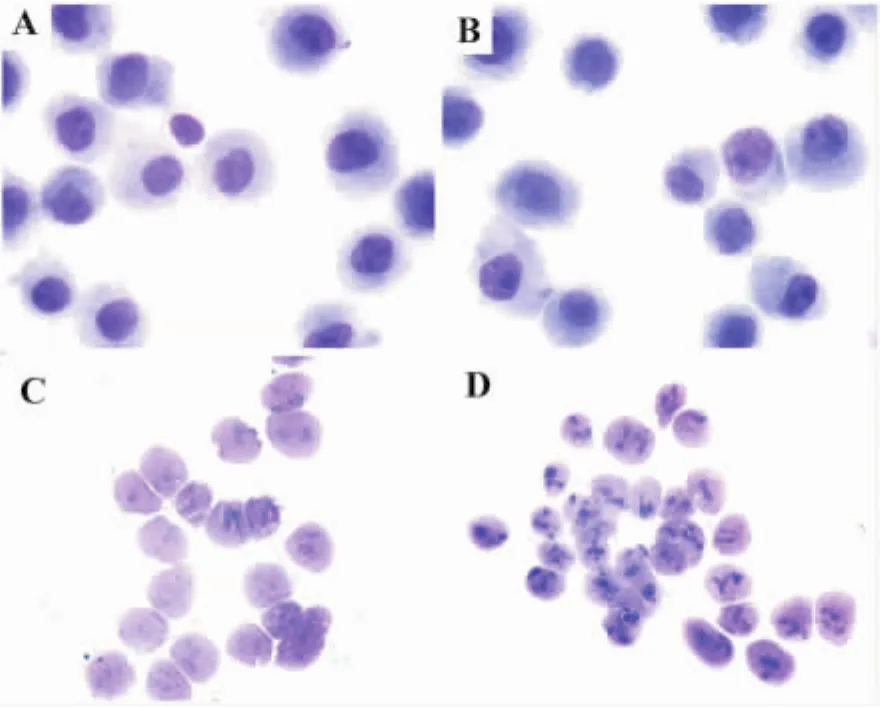
Fig.1 Profile of cells and isolated nuclei by light microscopy(×1000).Cells and isolated nuclei were both stained by May-Grünwald-Giemsa stain.Nuclei were stained pink and cytoplasm blue.A:Rho+cells;B:Rho0cells;C:Rho+cell nuclei;D:Rho0cell nuclei.
2.2 Nuclear proteome characters of Rho+and Rho0cells
The 2-DE patterns of nuclear extracts from Rho+and Rho0cells were exhibited in Fig.2 A.For 2-DE patterns of Rho+cell nuclei,a total of 125±5 protein-spots were detected and 121±8 protein-spots were matched with an average matching rate of 96.7%.For those of Rho0cell nuclei,a total of 126±11 proteinspots were detected and 122±14 protein-spots were matched with an average matching rate of 97.0%.The difference in nuclear protein expression between Rho+and Rho0cells was analyzed using PD Quest Version 8.0 software.Compared to Rho+cell nuclei,11 polypeptides down-regulated and 21 polypeptides up-regulated in Rho0cell nuclei(P<0.05).
To investigate the low relative molecular mass(<20 ku)protein profiles in Rho+and Rho0cell nuclei,SELDI-TOF ProteinChip technology was employed.As shown in Fig.2 B,4 protein-peaks having molecular mass around 6271.8,112 94.1,137 67.5 and 152 94.9 u significantly decreased in Rho0cell nuclei(P<0.05).
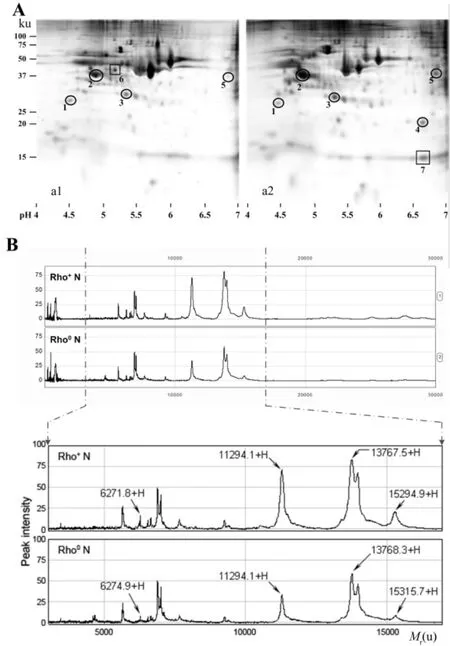
Fig.2 Nuclear proteome characters in Rho+and Rho0 cells.A:2-DE maps of Rho+(a1)and Rho0(a2)cell nuclei.Seven protein spots marked on the maps were differentially expressed and identified by MS.Five protein spots marked with circles were successfully identified as proteins,but other two protein spots marked with squares failed to be identified as proteins.B:SELDI-TOF MS profiles of Rho+and Rho0cell nucleus extracts on a NP20 ProteinChip.
2.3 Identification of differentially expressed proteins
Based on the analysis mentioned above,seven differentially expressed protein spots marked on 2-DE maps(Fig.2 A)were excised and analyzed with MALDI-TOF MS.Five of these protein spots(circled)shown in Fig.3 A and Fig.3 B were successfully identified as eukaryotic translation initiation factor 6(eIF-6),nucleophosmin(NPM),splicing factor,arginine/serine-rich 1(SFRS1),splicing factor,arginine/serine-rich 3(SFRS3)and heterogeneous nuclear ribonucleoprotein G(hnRNP G).These proteins were mostly located in nuclei,and participated in various cellular functions such as regulation of gene expression,RNA processing and ribosome biogenesis.The details of each identified protein were listed in Tab.1 .
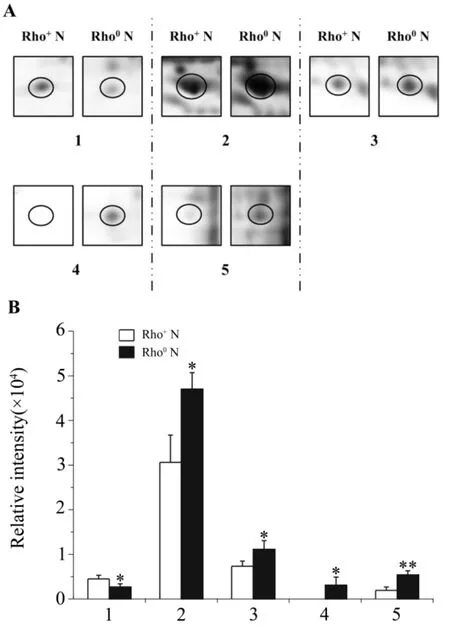
Fig.3 Magnified 2-DE maps(A)and relative intensity(B)of spots successfully identified by MALDI-TOF MS.Magnified 2-DE maps(A)and relative intensity(B)of spots marked with circles in Fig.2 A.1-5:differentially expressed proteins spots.±s,n=3.*P<0.05,**P<0.01,compared with Rho+N.
2.4 Confirmation of nucleophosmin expression levels in Rho+and Rho0cells
As shownin Fig.4 ,toconfirmthe proteomic analysis results,the cellular levels of nucleophosmin in Rho+and Rho0cells weredetermined by Western blotting.The cellular levels of nucleophosmin in Rho0cells were significantly higher than those in Rho+cells(P<0.01),which were consistent with the proteomic analysis results.

Tab.1 Characterization of differentially expressed proteins in Rho0and Rho+cell nuclei
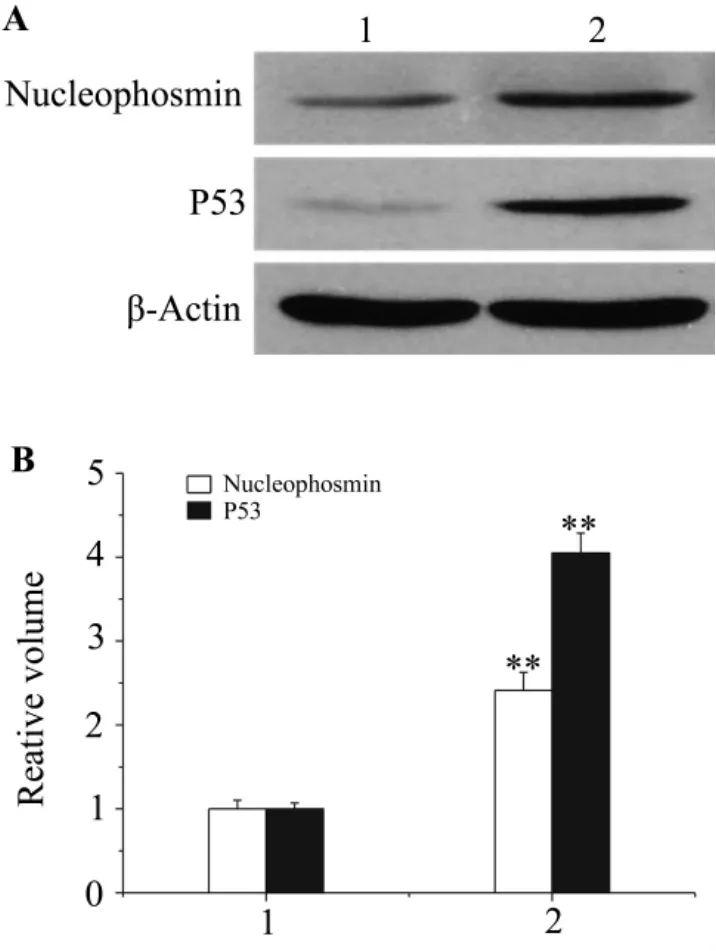
Fig.4 Expression of nucleophosmin and P53 in Rho+and Rho0cells.1:Rh+cells;2:Rh0cells.A:representative immunoblot obtained with anti-nucleophosmin or anti-P53 antibodies.B:densitometric analysis of A.The value in Rho+cells was determined to 1.±s,n=3.**P<0.01,compared with Rho0cells.
2.5 Expression of P53 and mitochondrial membrane potential(MMP)in Rho+and Rho0cells
Protein-protein association analysis of the identified proteins by STRING version 8.3 indicates that four proteins except eIF-6 were in a relevant molecular network and associated directly or indirectly with P53 which was reported to participate in MMP generation in mtDNA-depleted cells.Therefore,P53 expression levels and MMP in Rho+and Rho0cells were measured by Western blotting and laser scanning confocal microscopy,respectively.As shown in Fig.4 and Fig.5 ,P53 expression in Rho0cells was significantly higher than that in Rho+cells.However,the MMP was basically consistent in Rho+and Rho0cells.
3 DISCUSSION
In the present study,the proteome profiling of Rho0cells was found to be different from that of their parental cells based on 2-DE and SELDITOF Protein Chip analysis,and five differentially expressed proteins were identified by MALDI-TOF MS.According to function,the identified proteins could be classified into two categories:one was related to regulation of gene expression,and the other was involved in ribosome biogenesis.
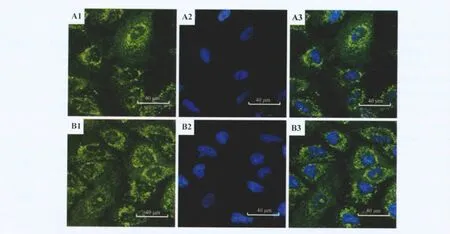
Fig.5 Mitochondrial membrane potential(MMP)in Rho0and Rho+cells.A:Rho+cells;B:Rho0cells.1:MMP;2:nucleus;3:merge.
NPM and eIF-6 are involved in ribosome biogenesis.NPM is a multifunctional protein with roles in various cellular processes,such as ribosome biosynthesis,centrosome duplication,protein chaperoning,histone assembly,cell proliferation,and regulation of tumor suppressors TP53/P53 and ARF[10-11].NPM regulates the increase in stability and transcriptional activation of P53 through inhibition of human MDM2,a nucleoplasmic and nucleolar RING-finger protein that acts as an E3 ubiquitin ligase initiating P53 proteasomal degradation[12-15].Therefore,the increased cellular levels of P53 in Rho0cells might be due to the up-regulation of NPM.P53 participates in regulating the expression of mitochondrial chloride intracellular channel(mtCLIC)protein that could be involved in mitochondrial membrane potential generation in mtDNA-depleted cells,a feature required to prevent apoptosis and to drive continuous protein import into mitochondria[16].The high-expression of NPM and P53 in Rho0cells might contribute to the maintenance of MMP in Rho0cells.Protein eIF-6 is an evolutionary conserved regulator of ribosomal function.It binds to the 60s ribosomal subunit and prevents its association with the 40s ribosomal subunit to form the 80s initiation complex.In addition,eIF-6 also contributes to the formation of nuclear matrix filaments[17].
hnRNP G,SFRS1 and SFRS3 contribute to the regulation of gene expression.hnRNP G plays an important role in RNA processing and metabolism[18-20]and it is detected primarily in the nuclei of mammalian cells and associated with nascent RNA transcripts as a part of transcriptional complexes[19].It alters pre-mRNA splicing pattern by antagonizing the effects of Tra2β,a splicing activator[21].Probably,its expression level may affect global gene expression and maintenance of normal cellular homeostasis[22].SFRS1 and SFRS3 belong to the splicing factor SR family.SFRS1 and SFRS3 are both involved in RNA processing in relation to cellular proliferation and/or maturation.In higher eukaryotes,pre-m RNA splicing is an essential step for expression of most protein-coding genes.Furthermore,alternative splicing of premRNA is used to regulate gene expression and to produce multiple proteins from a single gene[23].Arecent study estimates that at least 74%of human multiexon genes are alternatively spliced[24].So the up-regulation of SFRS1 and SFRS3 may be important to nuclear gene expression of Rho0cells.
In summary,mtDNA-depletion induced nuclear proteome alteration.Down-regulation of eIF-6and up-regulation of NPM,SFRS1,SFRS3 and hnRNP G in Rho0cell nuclei were found.These findings will shed light on the responses of nuclei to mitochondrial dysfunction.
[1]Chan DC.Mitochondria:dynamic organelles in disease,aging,and development[J].Cell,2006,125(7):1241-1252.
[2]Wallace DC.A mitochondrial paradigm of metabolic and degenerative diseases,aging,and cancer:a dawn for evolutionary medicine[J].Annu Rev Genet,2005,39:359-407.
[3]Cannino G,Di Liegro CM,Rinaldi AM.Nuclear-mitochondrial interaction[J].Mitochondrion,2007,7(6):359-366.
[4]Liu Z,Butow RA.Mitochondrial retrograde signaling[J].Annu Rev Genet,2006,40:159-185.
[5]Delsite R,Kachhap S,Anbazhagan R,Gabrielson E,Singh KK.Nuclear genes involved in mitochondria-tonucleus communication in breast cancer cells[J].Mol Cancer,2002,1:6.
[6]Henrich S,Cordwell SJ,Crossett B,Baker MS,Christopherson RI.The nuclear proteome and DNA-binding fraction of human Raji lymphoma cells[J].Biochim Biophys Acta,2007,1774(4):413-432.
[7]Zhao P,Zhong W,Ying X,Yao B,Yuan Z,Fu J,et al.Comparative proteomic analysis of anti-benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide-transformed and normal human bronchial epithelial G0/G1cells[J].Chem Biol Interact,2010,186(2):166-173.
[8]Hodgetts A,Bossé JT,Kroll JS,Langford PR.Analysis of differential protein expression in Actinobacillus pleuropneumoniae by surface enhanced laser desorption ionisation-ProteinChip(SELDI)technology[J].Vet Microbiol,2004,99(3-4):215-225.
[9]Zhang S,Fu J,Zhou Z.Changes in the brain mitochondrial proteome of male Sprague-Dawley rats treated with manganese chloride[J].Toxicol Appl Pharmacol,2005,202(1):13-17.
[10]Bergstralh DT,Conti BJ,Moore CB,Brickey WJ,Taxman DJ,Ting JP.Global functional analysis of nucleophosmin in Taxol response,cancer,chromatin regulation,and ribosomal DNA transcription[J].Exp Cell Res,2007,313(1):65-76.
[11]Okuda M.The role of nucleophosmin in centrosome duplication[J].Oncogene,2002,21(40):6170-6174.
[12]Colombo E,Marine JC,Danovi D,Falini B,Pelicci PG.Nucleophosmin regulates the stability and transcriptional activity of P53[J].Nat Cell Biol,2002,4(7):529-533.
[13]Coutts AS,Adams CJ,La Thangue NB.p53 ubiquitination by Mdm2:a never ending tail?[J].DNA Repair(Amst),2009,8(4):483-490.
[14]Kurki S,Peltonen K,Latonen L,Kiviharju TM,Ojala PM,Meek D,et al.Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation[J].Cancer Cell,2004,5(5):465-475.
[15]Wang QQ,Zhang ZY,Xiao JY,Yi C,Li LZ,Huang Y,et al.Knockdown of nucleophosmin induces S-phase arrest in HepG2 cells[J].Chin J Cancer,2011,30(12):853-860.
[16]Arnould T,Mercy L,Houbion A,Vankoningsloo S,Renard P,Pascal T,et al.mtCLIC is up-regulated and maintains a mitochondrial membrane potential in mtDNA-depleted L929 cells[J].FASEB J,2003,17(14):2145-2147.
[17]Ceci M,Offenhäuser N,Marchisio PC,Biffo S.Formation of nuclear matrix filaments by p27(BBP)/eIF6[J].Biochem Biophys Res Commun,2002,295(2):295-299.
[18]Dreyfuss G,Matunis MJ,Piñol-Roma S,Burd CG.hnRNP proteins and the biogenesis of mRNA[J].Annu Rev Biochem,1993,62:289-321.
[19]Soulard M,Della Valle V,Siomi MC,Piñol-Roma S,Codogno P,Bauvy C,et al.hnRNP G:sequence and characterization of a glycosylated RNA-binding protein[J].Nucleic Acids Res,1993,21(18):4210-4217.
[20]Soulard M,Barque JP,Della Valle V,Hernandez-Verdun D,Masson C,Danon F,et al.A novel 43-kDa glycoprotein is detected in the nucleus of mammalian cells by autoantibodies from dogs with autoimmune disorders[J].Exp Cell Res,1991,193(1):59-71.
[21]Nasim MT,Chernova TK,Chowdhury HM,Yue BG,Eperon IC.HnRNP G and Tra2beta:opposite effects on splicing matched by antagonism in RNA binding[J].Hum Mol Genet,2003,12(11):1337-1348.
[22]Shin KH,Kim RH,Kang MK,Kim RH,Kim SG,Lim PK,et al.p53 promotes the fidelity of DNA endjoining activity by,in part,enhancing the expression of heterogeneous nuclear ribonucleoprotein G[J].DNA Repair(Amst),2007,6(6):830-840.
[23]Shen H,Green MR.A pathway of sequential arginineserine-rich domain-splicing signal interactions during mammalian spliceosome assembly[J].Mol Cell,2004,16(3):363-373.
[24]Johnson JM,Castle J,Garrett-Engele P,Kan Z,Loerch PM,Armour CD,et al.Genome-wide survey of human alternative pre-mRNA splicing with exon juncteion microarrays[J].Science,2003,302(5653):2141-2144.
线粒体DNA缺失A549细胞与其母本细胞核蛋白质组差异分析
赵鹏1,张朝晖1,2,仲伟鉴3,应贤平3,袁准1,姚碧云1,傅娟玲1,周宗灿1
(1.北京大学医学部毒理学系,北京100191;2.南华大学环境医学和放射卫生院,湖南衡阳421001;3.上海市疾病预防控制中心毒理科,上海200336)
目的比较线粒体DNA(mtDNA)缺失A549细胞(Rho0细胞)与其母本细胞(Rho+细胞)核蛋白表达谱,并探讨细胞核对线粒体功能缺陷的应答反应。方法二维凝胶电泳(2-DE)和表面增强激光法解吸电离-飞行时间(SELDI-TOF)蛋白芯片测定Rho0细胞和Rho+细胞核蛋白表达谱,基质辅助激光解吸电离-飞行时间(MALDI-TOF)质谱结合数据库检索鉴定差异表达的蛋白点,Western印迹法测定核磷蛋白和P53表达,激光共聚焦显微镜测定线粒体膜电位。结果2-DE显示Rho0细胞核中11个蛋白点表达下调,21个蛋白点表达上调。基于NP20蛋白质芯片的SELDI-TOF质谱分析发现4个蛋白质峰在Rho0细胞核中明显下降。其中1个表达下调的蛋白点被鉴定为eIF-6,4个表达上调的蛋白点被鉴定为核磷蛋白,SFRS1,SFRS3和hnRNP G。Western印迹实验结果显示,Rho0细胞中核磷蛋白表达增加。P53和线粒体膜电位(MMP)测定结果显示,Rho0细胞中P53表达高于Rho+细胞,两种细胞MMP基本一致。结论mtDNA缺失诱导了细胞核蛋白质组改变。Rho0细胞可以作为研究线粒体与核交互作用的模型。
核蛋白质组;基因,p53;膜电位,线粒体
国家自然科学基金(30671781);国家自然科学基金(81001253);北京大学医学部新教师科研启动基金(BMU20090460)
周宗灿,E-mail:zhouzc@bjmu.edu.cn,Tel:(010)82801531
2011-06-01接受日期:2012-03-19)
R963
A
1000-3002(2012)04-0482-07
10.3867/j.issn.1000-3002.2012.04.003
The project supported by National Natural Science Foundation of China(30671781);National Natural Science Foundation of China(81001253);and Fundamental Research Funds for Central Universities(BMU20090460)
Biographies:ZHAO Peng(1979-),Ph.D.,main research field is toxicology.ZHOU Zong-can(1944-),professor,main research field is toxicology.
ZHOU Zong-can,Tel:(010)82801531,E-mail:zhouzc@bjmu.edu.cn
(本文编辑:付良青)

