Complete genome sequences of four isolates of Citrus leaf blotch virus from citrus in China
LI Ping, LI Min, ZHANG Song, WANG Jun, YANG Fang-yun CAO Meng-ji, LI Zhong-an
1 National Citrus Engineering Research Center, Citrus Research Institute, Southwest University, Chongqing 400712, P.R.China
2 College of Agronomy and Biotechnology, Southwest University, Chongqing 400715, P.R.China
1.Introduction
Citrus leaf blotch virus (CLBV) is a member of the genus Citrivirus in the family Betaflexiviridae (Adams et al.2012).It has been found in Spain, Japan, USA, France, Austrialia,Italy, New Zealand, Cuba, and China (Vives et al.2002b;Guardo et al.2007; Harper et al.2008; Hernández-Rodríguez et al.2016; Cao et al.2017).CLBV was first detected in Nagami Kumquat (Fortunella margarita Lour.Swingle) from Corsica, France (Navarro et al.1984) and later was found to associate with bud union disorder on Troyer citrange (Citrus sinensis×Poncirus trifoliata) (Galipienso et al.2001).It is a graft- and seedtransmissible pathogens, albeit at low percentages in citrus(Guerri et al.2004).This virus is able to infect a wide range of hosts including numerous fruits such as citrus, kiwi,sweet cherry and an ornamental plant peony (Galipienso et al.2001; Chavan et al.2013; Wang et al.2016; Gress et al.2017), as well as several herbaceous hosts, such as Nicotiana occidentails, Nicotiana benthamiana, Nicotiana cavicola, Nicotiana glutinosa, and Nicotiana clevelandii included (Vives et al.2008; Guardo et al.2009; Chavan et al.2013).In China, CLBV was initially found in kiwi,and then in sweet cherry and citrus (Wang et al.2016; Zhu et al.2016; Cao et al.2017).
The virus particle of CLBV is filamentous and flexuous,960 nm long and 14 nm in diameter.The genome of CLBV is a single-stranded positive-sense RNA with full length of approximately 8.7 kb, excluding a 3´-terminal poly(A) tail, and comprises three overlapping open reading frames (ORFs).ORF1 encodes a polyprotein with a molecular mass of about 227.4 kDa which contains the viral replication components with methyl-transferase, AlkB-like, OTu-like peptidase, papain-like protease, helicase and RNA-dependent RNA polymerase (RdRp) motifs.ORF2 encodes a 40.2-kDa putative cell-to-cell movement protein (MP).ORF3 codes an approximately 40.7-kDa coat protein (CP) (Vives et al.2001).In previous studies,CLBV produced two 3´-coterminal and two 5´-coterminal subgenomic RNAs which were essential for the viral infection (Vives et al.2002a).In this study, the complete nucleotide sequences of CLBV isolates from different citrus cultivars and regions were first obtained in China.This study would provide the basis for further studies on molecular evolution of CLBV.
2.Materials and methods
2.1.Virus isolates and RNA extraction
For this research, a total of 289 citrus samples were collected amongst different cultivars from several major citrus-growing regions of China including Zhejiang,Hunan, Sichuan, Jiangxi, Yunnan, and Chongqing.Total RNA extracts were obtained from the leaf tissues using TRIzol®Reagent (Thermo-Fisher, USA) according to the manufacturer’s instructions, and then one-step RT-PCR detection with primer pair CLBV-F (5´-AGC CATAGTTGAACCATTCCTC-3´) and CLBV-R (5´-GC AGATCATTCACCACATGC-3´) (Harper et al.2008) was used.The four CLBV-infected samples featured with different cultivars and geographical locations were selected for amplification of the complete virus genome.
2.2.RT-PCR and genome cloning
In order to obtain full genomic sequence, specific primers were initially designed into overlapping fragments based on the sequences of a CLBV isolate from GenBank (accession no.FJ009367) (Table 1).For the determination of the 5´- and 3´-terminal sequences of the genomic RNA, a commercial RACE Kit (Invitrogen, USA) was used.All fragments were amplified using the PreimeScriptTMOne-Step RT-PCR Kit version 2 (TaKaRa, China).PCR amplification was performed in a total reaction volume of 50 μL with 1.25 μL each primer in pair (10 μmol L-1) according to the reagent manuals.The thermal cycling conditions for RT-PCR were one cycle of 45°C for 30 min and 94°C for 3 min, 40 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 150-180 s (adapt for amplicon length), and a final elongation at 72°C for 10 min.
PCR products of given size were verified and purified using a gel DNA extraction kit (OMEGA Bio-Tek, USA).These final amplicons were cloned into pEASY-T1 vector(TransGen Biotech, China), and five clones per fragment were custom sequenced.
2.3.Sequence analysis
The clones derived from six overlapping fragments were assembled with classic option and the outcome sequences were modified and decided (including 5´- and 3´-RACE) by DNASTAR 7 (DNASTAR Inc., USA).The genomic sequence and both nucleotides and amino acids (aa) sequence of ORF1, ORF2 and ORFE3 were separately compared with other available CLBV isolates in GenBank by CLC Main Workbench 7.9 (Qiagen, German).The phylogenetic tree was constructed by the Clustal-W and neighbor-joining method with 1 000 bootstrap replications using the MEGA version 7.0 Program.
3.Results
A total of 15 citrus samples were detected to be CLBV-positives by RT-PCR and were divided into four classes by sequence analysis, which were collected from four different cultivars Reikou in Sichuan (LH), Yura Wase in Zhejiang (YL), Bingtangcheng in Hunan (BT) and Fengjie 72-1 in Chongqing (FJ), respectively.Complete genome sequence of a typical isolate for each class was deposited in GenBank database under accession number MF784853 (CLBV-LH) (Appendix A), MF784854 (CLBV-YL)(Appendix B), MF784855 (CLBV-BT) (Appendix C) and MF784856 (CLBV-FJ) (Appendix D).The complete genome of all the four CLBV isolates were 8 747 nucleotides (nt) long,excluding the poly(A) tail.The isolates were 73.7-98.1%identical to other available CLBV isolates from GenBank.Analysis of the genomic structures showed that all the four isolates were similar to previously reported CLBV citrus isolate (AJ318061) (Vives et al.2001), and CLBV-LH, CLBVYL, CLBV-BT, and CLBV-FJ genomes shared 97.9, 97.8,98.0, and 98.0% identity, respectively, with AJ318061 isolate.These four isolates all contained two untranslated regions(UTR) of 73 and 541 nt at the 5´ and 3´ ends, respectively.The ORF1 began at nucleotide position 74 and ended at position 5 962, encoded a polypeptide of 1 962 aa.ORF2(5 962-7 050 nt) encoded a MP that was 362 aa.ORF3(7 115-8 206 nt) coded a 363-aa CP.When ORF1, ORF2,and ORF3 of the four isolates were compared with the database of NCBI, the nucleotide similarities were 71.7-98%, 77.4-97.8%, and 77.5-98.6%, respectively, and the amino acid similarities were 78.3-98.7%, 95.9-99.5%,and 88.2-99.7%, respectively.Sequence alignments demonstrated that CP and MP regions of CLBV isolates were both highly conserved in either citrus or kiwi in respects of alignments of whatever nucleotide or amino acid.
To reveal phylogenetic relationships, a tree was carried out based on the genomic sequences of the four CLBV isolates and nine others in the NCBI database.CLBV isolates from citrus in China were clustered in the same clade within the phylogenetic tree, and were closely related to the New Zealand isolate (EU857540) (Fig.1).
4.Discussion
CLBV was first detected in kiwi plants with a rate of 13.3%which grown in China (Zhu et al.2016).Subsequently, CLBV was found in sweet cherry and citrus with a rate of 15% in China (Wang et al.2016; Cao et al.2017).In contrast, our result showed a lower infected rate of 5.2% in citrus samples likely due to random sampling, while the statistics covers still limited for only parts of regions were investigated and only four cultivars were found CLBV-infected.Generally,complete genome sequence of CLBV for Chinese isolates were only reported in sweet cherry and kiw.In our study,four genome sequences of CLBV isolated from citrus were obtained, of which CLBV-LH and CLBV-YL, CLBV-BT and CLBV-FJ were identified as Citrus reticulata (L.) and Citrus sinensis (L.), respectively.The four isolates shared high resemblance to each other and homologous with those from other countries, indicating low genetic variation within citrus groups of CLBV isolates from whatever geographical origins,as previously reported (Vives et al.2002b).
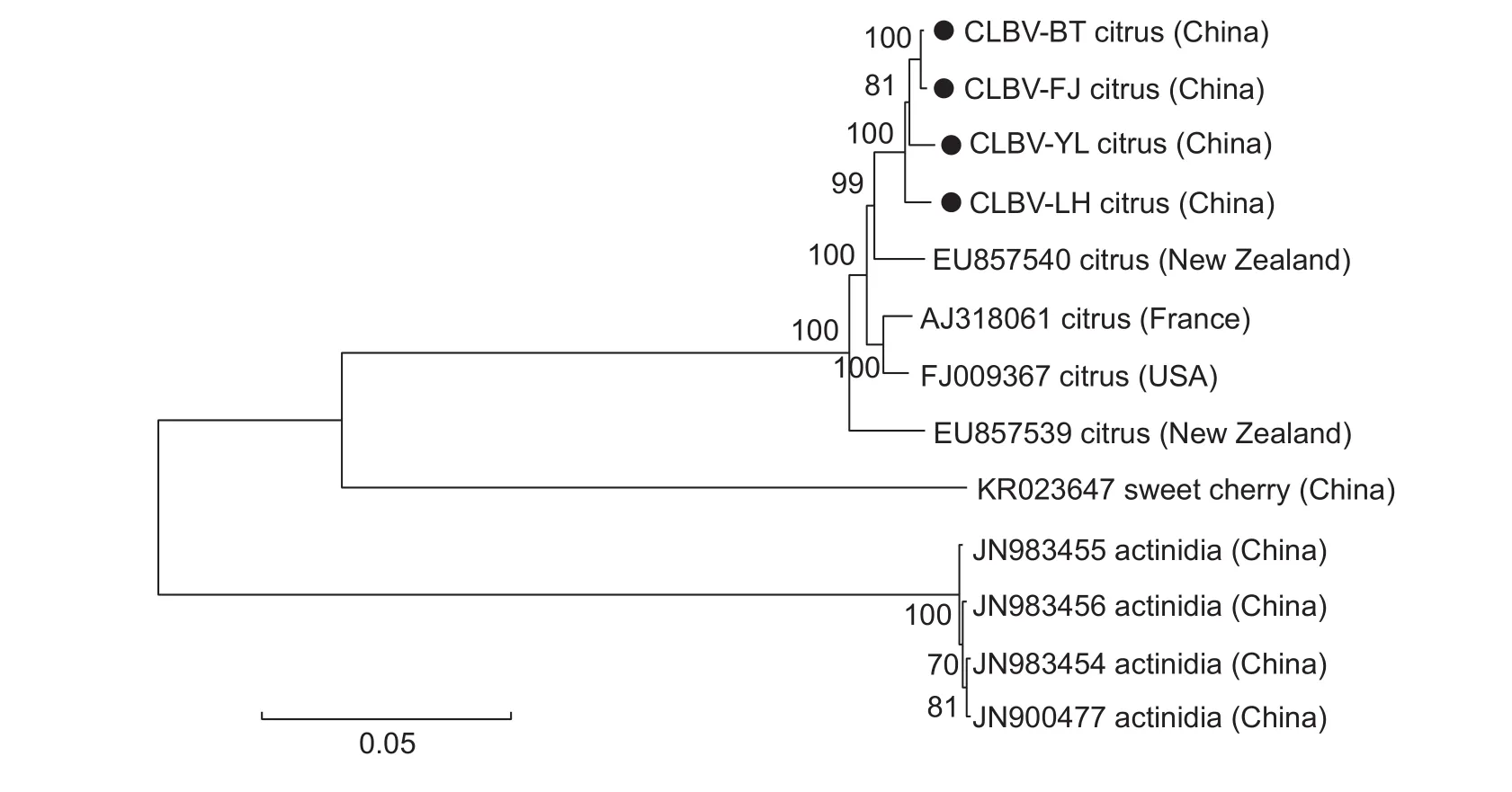
Fig.1 Phylogenetic tree based on the genome sequences of the four Citrus leaf blotch virus (CLBV) isolates in this study and other available CLBV isolates from GenBank.The tree was constructed by the neighbor-joining method with 1 000 bootstrap replicates.The four CLBV genome sequences from this study are indicated by “”, and the accession number, host and country of each isolate were indicated.
5.Conclusion
To our knowledge, this study provides the first complete nucleotide sequences of CLBV isolates from citrus in China,therefore enriches bioinformatics database.The spreading and severity of CLBV could be determined by the surveys in six regions of China.These mentioned details would form the basis for development of molecular diagnostic and hence a more effective disease control strategy.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31501611), the Chongqing Research Program of Basic Research and Frontier Technology,China (cstc2017jcyjBX0016), the Chongqing Science and Technology Commission Project, China (cstc2016shmsztzx80003), the Fundamental Research Funds for the Central Universities, China (XDJK2016B21, SWU116012), and the Special Fund for Agro-scientific Research in the Public Interest, China (201203076-01).
Appendicesassociated with this paper can be available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
Adams M J, Candresse T, Hammond J, Kreuze J F, Martelli G P,Namba S, Pearson M N, Ryu K H, Saldarelli P, Yoshikawa N.2012.Family Betaflexiviridae.In: King A M Q, Adams M J, Carstens E B, Lefkowitz E J, eds., Virus Taxonomy:Ninth Report of the International Committee on Taxonomy of Viruses.Elsevier Academic Press, London.pp.920-941.
Cao M J, Yu Y Q, Tian X, Yang F Y, Li R H, Zhou C Y.2017.First report of Citrus leaf blotch virus in lemon in China.Plant Disease, 101, 1561.
Chavan R R, Blouin A G, Cohen D, Pearson M N.2013.Characterization of the complete genome of a novel citrivirus infecting Actinidia chinensis.Archives of Virology, 158,1679-1686.
Galipienso L, Vives M C, Moreno P, Milne R G, Navarro L, Guerri J.2001.Partial characterisation of Citrus leaf blotch virus,a new virus from Nagami kumquat.Archives of Virology,146, 357-368.
Guardo M, Potere O, Castellano M A, Savino V, Caruso A.2009.A new herbaceous host of Citrus leaf blotch virus.Journal of Plant Pathology, 91, 485-488.
Guardo M, Sorrentino G, Marletta T, Caruso A.2007.First report of Citrus leaf blotch virus on Kumquat in Italy.Plant Disease, 91, 1054.
Guerri J, Pina J A, Vives M C, Navarro L, Moreno P.2004.Seed transmission of Citrus leaf botch virus: Implications in quarantine and certification programs.Plant Disease,88, 906.
Gress J C, Smith S, Tzanetakis I E.2017.First report of Citrus leaf blotch virus in peony in the USA.Plant Disease, 101,637.
Harper S, Chooi K, Pearson M.2008.First report of Citrus leaf blotch virus in New Zealand.Plant Disease, 92, 1470.
Hernández-Rodríguez L, Pérez-Castro J M, García-García G, Ramos-González P L, Zamora-Rodríguez V, Ferriol-Marchena X, Peña-Bárzaga I, Riverend B L.2016.Citrus leaf blotch virus in Cuba: First report and partial molecular characterization.Tropical Plant Pathology, 41, 147-154.
Navarro L, Pina J, Ballester-Olmos J, Moreno P, Cambra M.1984.A new graft transmissible disease found in Nagami kumquat.In: Timmer L W, ed, Proceedings of the 9th Conference International Organisation of Citrus Virologists.IOCV Riverside, CA.pp.234-240.
Vives M C, Galipienso L, Navarro L, Moreno P, Guerri J.2001.The nucleotide sequence and genomic organization of Citrus leaf blotch virus: Candidate type species for a new virus genus.Virology, 287, 225-233.
Vives M C, Galipienso L, Navarro L, Moreno P, Guerri J.2002a.Characterization of two kinds of subgenomic RNAs produced by Citrus leaf blotch virus.Virology, 295, 328-336.
Vives M C, Martin S, Ambr O S S, Renovell A, Navarro L, Pina J A, Moreno P, Guerri J.2008.Development of a full-genome cDNA clone of Citrus leaf blotch virus and infection of citrus plants.Molecular Plant Pathology, 9, 787-797.
Vives M C, Rubio L, Galipienso L, Navarro L, Moreno P, Guerri J.2002b.Low genetic variation between isolates of Citrus leaf blotch virus from different host species and of different geographical origins.Journal of General Virology, 83, 2587.
Wang J, Zhu D, Tan Y, Zong X, Wei H, Liu Q.2016.First report of Citrus leaf blotch virus in sweet cherry.Plant Disease,100, 1027.
Zhu C X, Wang G P, Zheng Y Z, Yang Z K, Wang L P, Xu W X,Hong N.2016.RT-PCR detection and sequence analysis of coat protein gene of Citrus leaf blotch virus infecting kiwifruit trees.Acta Phytopathologica Sinica, 46, 11-16.(in Chinese)
 Journal of Integrative Agriculture2018年3期
Journal of Integrative Agriculture2018年3期
- Journal of Integrative Agriculture的其它文章
- Characteristic analysis of tetra-resistant genetically modified rice
- A wheat gene TaSAP17-D encoding an AN1/AN1 zinc finger protein improves salt stress tolerance in transgenic Arabidopsis
- Characterization of GhSERK2 and its expression associated with somatic embryogenesis and hormones level in Upland cotton
- GmNAC15 overexpression in hairy roots enhances salt tolerance in soybean
- Molecular cloning and functional characterization of a soybean GmGMP1 gene reveals its involvement in ascorbic acid biosynthesis and multiple abiotic stress tolerance in transgenic plants
- Responses of the antioxidant system to fluroxypyr in foxtail millet(Setaria italica L.) at the seedling stage
