A re-investigation of the bloom-forming unarmored dinoflagellate Karenia longicanalis (syn. Karenia umbella)from Chinese coastal waters*
WANG Jianyan (王建艳) , CEN Jingyi (岑竞仪) , LI Si (李思) ,LÜ Songhui (吕颂辉) , , MOESTRUP Øjvind , CHAN Kin-Ka (陈健嘉),JIANG Tao (江涛) , LEI Xiangdong (雷向东)
1 Department of Science Research, Beijing Museum of Natural History, Beijing 100050, China
2 College of Life Science and Technology, Jinan University, Guangzhou 510632, China
3 Key Laboratory of Eutrophication and Red Tide Prevention of Guangdong Higher Education Institutes, Jinan University,Guangzhou 510632, China
4 Marine Biological Section, Department of Biology, University of Copenhagen, Universitetsparken 4, DK-2100 Copenhagen Ø,Denmark
5 School of Science & Technology, Open University of Hong Kong, 999077 Kowloon, Hong Kong, China
6 Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China
7 Wenzhou Marine Environmental Monitoring Center Station, State Oceanic Administration, Wenzhou 325013, China
Abstract The dinoflagellate genus Karenia is known for recurrent harmful blooms worldwide. However,species diversity of the genus is generally overlooked owing to the difficulty of identifying small unarmored dinoflagellates. We have established four clonal cultures of Karenia longicanalis isolated from the type locality, Hong Kong harbor (strain HK01) and other three locations along the Chinese coasts (strains YB01,DT01, and NJ01). The morphology of the strain was studied by light and scanning electron microscopy (LM and SEM) and the pigment composition analyzed by high-performance liquid chromatography. We provide the first molecular data of K. longicanalis based on the large subunit (LSU) rRNA gene sequence and internal transcribed spacer (ITS). The four strains showed identical LSU rDNA sequences with a similarity of 99.4% to the holotype of Karenia umbella (strain KUTN05) from Australia. In the ITS phylogeny, the sequence of K. umbella branched between the Chinese strains of K. longicanalis. A careful comparison of the morphology of K. longicanalis and K. umbella reveals the similarity in the diagnostic characters.Differences may appear due to the sample treatment for SEM. We conclude that K. umbella is a junior synonym of K. longicanalis.
Keyword: harmful algal blooms (HABs); phytoplankton; morphology; phylogeny; large subunit (LSU)rRNA; internal transcribed spacer (ITS)
1 INTRODUCTION
The Kareniaceae has been recognized as a separate family, different from the large polyphyletic generaGymnodiniumF. Stein andGyrodiniumKofoid &Swezy (Bergholtz et al., 2005). The family was erected to encompass the three gymnodinioid dinoflagellate generaKareniaGert Hansen &Moestrup,KarlodiniumJ. Larsen andTakayamade Salas, Bolch, Botes & Hallegraeff . Species of Kareniaceae are characterized by a straight or sigmoid apical groove on the epicone. Another key feature of this family is pigments of the chloroplasts, fucoxanthin or fucoxanthin-derivatives being the main accessory pigments (Daugbjerg et al., 2000; de Salas et al., 2003;Bergholtz et al., 2005). Gómez et al. (2005) suggested the phylogenetic relationship betweenBrachidiniumF. J.R. Taylor andKareniathat was later confirmed by molecular data (Henrichs et al., 2011).
Species ofKareniaare well known to form recurrent blooms in many parts of the world. Many species are responsible for harmful algal bloom events such as large-scale fish kills, other marine mortality,neurotoxic shellfish poisoning (NSP) and even respiratory problems in humans (Chang, 1999; Tester et al., 2000; Yang et al., 2000; Kempton et al., 2002;Botes et al., 2003; de Salas et al., 2004a, b; Naar et al.,2007; Hoagland et al., 2009 etc.), and most species of the genus may turn out to be toxic (Brand et al., 2012).Further studies have revealed thatKareniaspecies can produce a variety of toxins, e.g. brevetoxin inKareniabrevis(C.C. Davis) Gert Hansen & Moestrup(Lin et al., 1981; Shimizu et al., 1986), gymnocin inKareniamikimotoi(Miyake & Kominami ex Oda)Gert Hansen & Moestrup (Satake et al., 2002, 2005),gymnodimine inKareniaselliformisHaywood,Steidinger & L. MacKenzie (Seki et al., 1995; Miles et al., 2003), brevisulcatic acids inKarenia brevisulcata(F. H. Chang) Gert Hansen & Moestrup(Holland et al., 2012), polyether brevetoxin-2 (PbTx-2) inKareniapapilionaceaHaywood & Steidinger(Fowler et al., 2015) and toxic sterols or polyunsaturated fatty acids (PUFAs) (Mooney et al., 2007).
Accurate identification ofKareniaspecies is based on analysis of cell characteristics, such as overall shape of cell, cell length and width, dorsoventral flatness of the cell, the shape and the length of the apical groove, the length of the cingular displacement,presence or absence of the sulcal intrusion, shape and location of the cell nucleus, chloroplast number and distribution in the cell, etc. (Daugbjerg et al., 2000;Hansen et al., 2000; Yang et al., 2000; Botes et al.,2003; Haywood et al., 2004; de Salas et al., 2004a, b).WhenKareniaspecies are present in low numbers in field samples, they are readily overlooked, and special attentions are mainly paid when they attain high concentrations or form a bloom. The species may be difficult to identify owing to their small size(approximately 20 μm in length and 10 μm in width)and the naked cells, which can be easily deformed in fixatives, e.g. Lugol’s solution or formaldehyde. It is generally only after the species have been isolated and cultured in the lab that its toxic potential can be determined. In addition, someKareniaspecies are probably mixotrophic (Siano et al., 2009) and hard to culture, and the lacking of strains in culture further complicates species identification.
Thirteen species ofKareniahave been described(Gómez, 2012). In the Chinese coastal waters, seven species have been detected (K.brevis,K.brevisulcata(F. H. Chang) Gert Hansen & Moestrup,K.digitataZ.B. Yang, H. Takayama, Matsuoka & Hodgkiss,K.mikimotoi,K.longicanalisZ. B. Yang, Hodgkiss &Gert Hansen,K.bicuneiformisBotes, Sym & G. C.Pitcher, andK.papilionacea) (Liu, 2008; Law and Lee, 2013).
In this study, aKareniaspecies was isolated from the East and South China Sea. Based on light and electron microscopy, pigment composition, and rDNA sequences (partial LSU and ITS), we identified it asKarenialongicanalisthat was originally described from Hong Kong harbor after a bloom in 1998 (Yang et al., 2001). We compared the morphology and molecular data ofK.longicanalisandK.umbellade Salas, Bolch & Hallegraeff (de Salas et al., 2004a),and we concluded that these taxa are conspecific with priority forK.longicanalis.
2 MATERIAL AND METHOD
2.1 Sampling and culturing
Karenialongicanalisstrains DT01 and NJ01 were established by isolating single cells from seawater samples collected at Dongtou Island (27°51′29″N,121°09′09″E) and Nanji Island (27°27′22″N,121°06′00″E) in the East China Sea on 25 June 2016.Strain YB01 was from seawater samples collected from Dongyu Pier (22°33′31″N, 114°32′22″E), Daya Bay off the South China Sea on 29 April 2016. The three clonal cultures DT01, NJ01, and YB01 were all collected from field samples. Strain HK01 was isolated from Yim Tin Sai, Tolo Harbour of Hong Kong (22°27′14″N, 114°12′50″E) during aKareniabloom event in January 2016. All the cultures were maintained at 20°C in L1 medium (Guillard and Hargraves, 1993) at a salinity of 30, using an irradiance of 50 μmol photons/(m2·s) in a 12 h:12 h light:dark cycle, and deposited at the Research Center for Harmful Algae and Marine Biology, Jinan University, Guangzhou, China. The culture HK01 was lost after the molecular analyses.
2.2 Light microscopy (LM)
Cells were examined under an Olympus BX61 microscope (Olympus, Tokyo, Japan). Light micrographs were captured using a QImaging Retiga 4000R digital camera (QImaging, Surrey, BC,Canada). Cells (more than 20 individuals) in exponential phase were photographed under the microscope at ×400 magnification, and the cell length and width were measured using Image-Pro Plus 6.0 image acquisition and analysis software(QImaging, Surrey, BC, Canada). For epifluorescence micrographs, cultured cells in exponential phase were stained by adding an equal volume of 4’,6-diamidino-2-phenylindole dihydrochloride(DAPI) (Sangon Biotech, Shanghai, China) at a concentration of 5 μg/mL, and incubation in the dark at room temperature for 5 min. Cells were photographed using an Olympus BX61 microscope fitted with 360–370 nm excitation and 420–460 nm emission filters.
2.3 Scanning electron microscopy (SEM)
Three different fixation methods were used. In the first procedure, healthy cultured cells were fixed at 4°C overnight in glutaraldehyde at a final concentration of 2.5%, and then filtered through nuclepore filters with a pore size of 3 μm (Whatman,Little Chalfont, UK), followed by dehydration in an ethanol series of 10%, 30%, 50%, 70%, 90% and 95%, 5 min in each change, and finally in 100% for 10 min (two changes). In the second procedure,healthy cultured cells were fixed with an equal volume of 4% OsO4(prepared with filtered seawater)for 1 h at room temperature. The fixed culture was rinsed once in seawater and once in deionized water.The cells were dehydrated in an ethanol series of 10%, 30%, 50%, 70% and 90%, this time 10 min in each change, followed by two changes of 100% for 15 min. In the third procedure, field seawater samples were preserved in Lugol’s solution in situ (final concentration was about 1%) and post fixed at 4°C overnight in glutaraldehyde at a final concentration of 2.5%. Cells were dehydrated as in the first procedure. All dehydrated samples were criticalpoint dried using a Leica EM CPD300 Critical Point Dryer (Leica Microsystems, Mannheim, Germany),and the preparations subsequently sputter-coated with gold. Cells were observed in a field emission scanning electron microscope (FEI QUANTA 200F,Hillsboro, USA).
2.4 Pigment analysis
Ten milliliter algal culture in mid exponential phase was filtered onto 0.45 μm pore size Whatman GF/F glass fibre filters (Whatman, Maidstone, UK)and stored in a refrigerator at -80°C for later treatment.Pigment extraction and analysis were performed according to the methods described in Zapata et al.(2000). The filters were cut into pieces, transferred to a centrifuge, and then extracted with 3 mL 95%methanol in a sonication bath containing ice and water for 5 min under low light. The extract was filtered through a Teflon film with a pore size of 0.2 μm to remove cellular debris. One milliliter of the extract was mixed with 0.2 mL Milli-Q water, and 100 μL aliquot of the mixture was analyzed using reverse-phase HPLC. The HPLC system Agilent 1200 series is equipped with an auto sampler and diodearray detector (Model G1315C; Agilent Technologies Inc., Palo Alto, CA, USA). Pigments extracted were separated by a C8 column (Waters Symmetry,150 mm×4.6 mm, 3.5 μm particle size) thermostated at 25°C.
Pigments were identified and quantified using the following 23 pure pigment standards (DHI Inc,Copenhagen, Denmark): chlorophylla(Chla),chlorophyllb(Chlb), chlorophyllc2 (Chlc2),chlorophyllc3 (Chlc3), fucoxanthin (Fuco),diadinoxanthin (Dia-dino), peridinin (Peri),violaxanthin (Viola), alloxanthin (Allo), diatoxanthin(Diato), β, β-carotene (β, β-Car), prasinoxanthin(Pras), lutein (Lut), neoxanthin (Neo), zeaxanthin(Zea), 19’-hexanoyloxy-fucoxanthin (Hex-fuco),19’-butanoyloxy-fucoxanthin (But-fuco),pheophorbidea(Pheidea), canthaxanthin (Cantha),divinyl chlorophyll a (DV-Chla), pheophytina(Phea), Mg-2,4-divinylpheoporphyrin (MgDVP), and gyroxanthin diester (Gyr-de).
2.5 DNA extraction and amplification of LSU and ITS
About 10 mL of cell culture in exponential growth was collected by centrifugation, and total genomic DNA was extracted using a Takara MiniBEST Universal Genomic DNA Extraction Kit (TaKaRa,Dalian, China) according to the manufacturer’s protocol. LSU rDNA (D1-D3 region) and ITS regions were amplified using primers D1R (Scholin et al.,1994) and D3B (Nunn et al., 1996) and primers ITS1 and ITS4 (White et al., 1990). The temperature profile for the PCR amplification was 94°C for 3 min followed by 38 cycles of 94°C for 30 s, 57°C for 30 s,and 72°C for 1 min, with an extension at 72°C for 6 min. PCR products were purified and sent to Beijing Genomics Institute (BGI, Guangzhou, China) for sequencing.
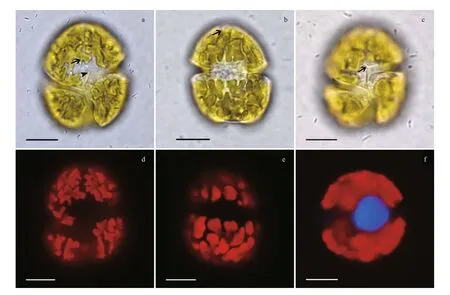
Fig.1 Light micrographs of Karenia longicanalis (strain YB01)
2.6 Sequence alignment and phylogenetic analysis
Sequences similar to our LSU rDNA and ITS sequences were identified using the BLAST server(https://blast.ncbi.nlm.nih.gov) and downloaded from GenBank. Details of sequences used for phylogenetic analysis were shown in Table S1 and Table S2.Sequences for phylogenetic analysis were aligned using ClustalX (Thompson et al., 1997). Sequence distances (percent identity and divergence) were calculated with the Megalign program within the DNASTAR sequence analysis package. Phylogenetic analyses were conducted using the Maximumlikelihood (ML) and Bayesian inference (BI) analyses.ML analysis was performed using MEGA version 6.0(Tamura et al., 2013) with the Kimura 2-parameter model and supported by 1 000 bootstrap replicates.Bayesian analyses were done using Mrbayes 3.1.2 following the best-fitting model (Ronquist and Huelsenbeck, 2003) and the best-fit model selected by MrModelTest 2.2 were SYM + G and GTR + G for LSU and ITS data set, respectively. The confidence limits were estimated using bootstrap analysis with 1 000 replications.
3 RESULT
3.1 Morphology of Karenia longicanalis
Karenialongicanalisis a relatively large-sized species compared to otherKareniaspecies. The cells are slightly dorsoventrally flattened. Length, width and length-width ratio of the three strains (YB01,DT01 and NJ01) are shown in Table 1. Cells have a conical epicone and an asymmetric hypocone(Fig.1a–c). The hypocone is truncated, the right lobe longer than the left lobe (Fig.1b, c). The apical groove is straight and very long, and it is clearly visible in the LM (Fig.1a). The cingulum is incised and displaced about 20% of the total cell length (Fig.1c). The sulcus is wide in the hypocone but becomes narrow in the inter-cingular region, and extends as a finger-like intrusion onto the epicone (Fig.1c). The cell possesses about 20 chloroplasts. Chloroplasts are bright yellowgreen, with many lobes (Fig.1d, e). The nucleus is round and centrally located close to the cell’s dorsal side (Fig.1f).
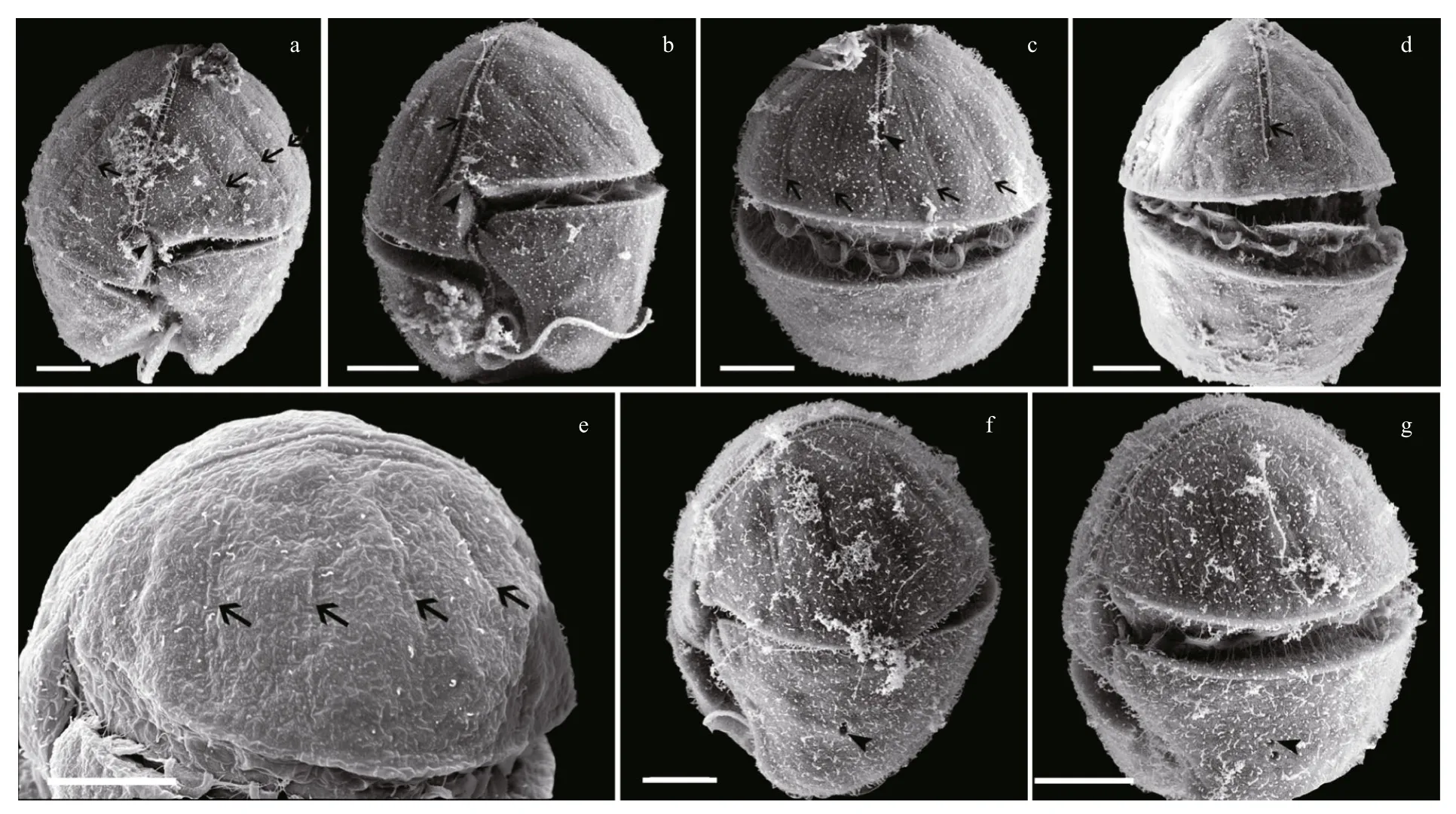
Fig.2 Scanning electron micrographs of Karenia longicanalis (strain YB01, fixed in OsO 4)
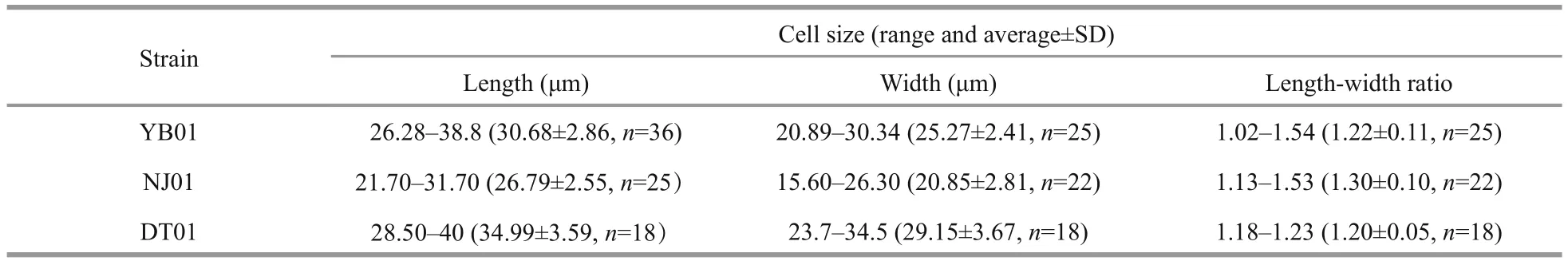
Table 1 Cell size of the three Karenia longicanalis strains isolated in this study
Under SEM, eight furrows are seen to radiate from the cell apex to the upper margin of the cingulum(Fig.2a, b, c, e). The apical groove is clearly observed extending down the ventral and dorsal sides of the epicone (Fig.2a–d), on the ventral side extending below the upper section of the cingulum (Fig.2b). The sulcal intrusion onto the epicone is short (Fig.2a, b). On the dorsal side, the apical groove extends about 1/2–2/3 or even 3/4 down the epicone (Fig.2c, d). It is shallow and less than 1 μm wide. One or more lateral pores are sometimes present on the ventral part of the hypocone,and usually on the left lobe of the hypocone (Fig.2f, g).Samples fixed in glutaraldehyde (Fig.3)were a little more swollen compared to samples fixed in OsO4.Lateral pores were not observed in the glutaraldehyde fixation and the furrows on the epicone were hard to see (Fig.3). Under SEM observations, the Lugolpreserved cells showed clearly a long apical groove,which extended near the upper cingulum (Fig.4a–c).Striae were usually not visible. The sulcal intrusion was short and sometimes could not be observed(Fig.4c). Lateral pores were regularly observed on the left lobe of the hypocone, numbering from one to four and clustering in different shapes (Fig.4d, e).
3.2 Pigments composition of Karenia longicanalis
A HPLC profile ofK.longicanalis(strain YB01) is shown in Fig.5. Five chlorophylls, Chla, Chlc2, Chlc3, pheophorbideaand MgDVP were detected. Eight carotenoids were also detected, of which fucoxanthin was the most abundant. The abundance of chlorophyll and carotenoid pigments expressed as mass ratios with respect to Chlaare shown in Table 2. The chromatogram (Fig.5) is very similar to previous reports onK.umbella, with some differences in the concentration of pigments.
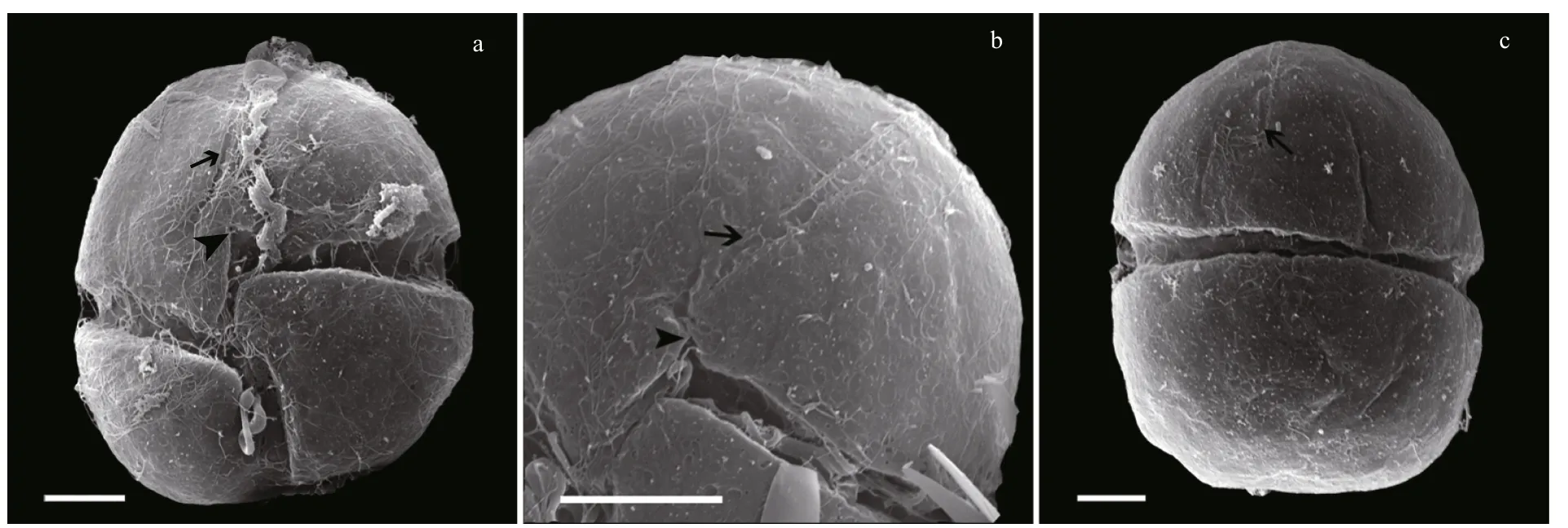
Fig.3 Scanning electron micrographs of Karenia longicanalis (strain YB01, fixed in glutaraldehyde)

Fig.4 Scanning electron micrographs of Karenia longicanalis (field samples, preserved in Lugol’s solution in situ and post fixed in glutaraldehyde)
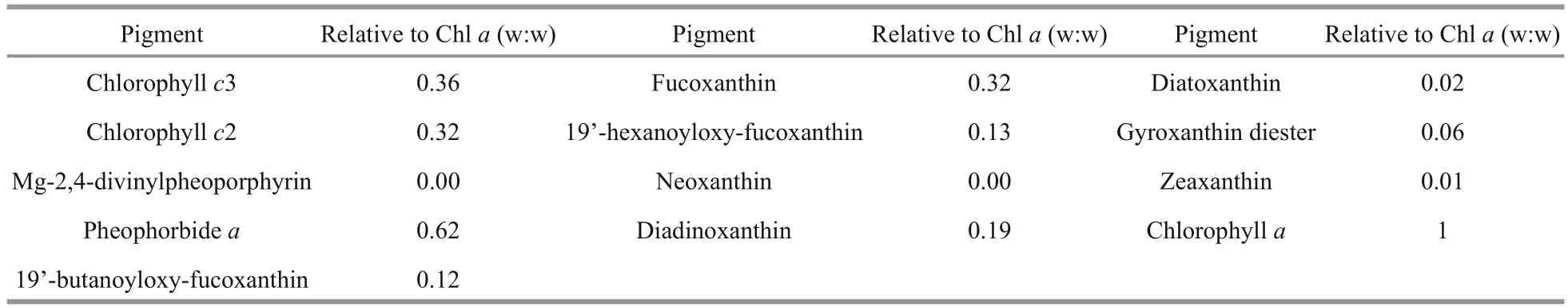
Table 2 Summary of HPLC pigment composition of K ar e nia longicanalis and their abundance (abundance of pigments is expressed in proportion to Chl a

Fig.5 Pigment chromatogram of Karenia longicanalis (strain YB01) from Daya Bay, Guangdong, China
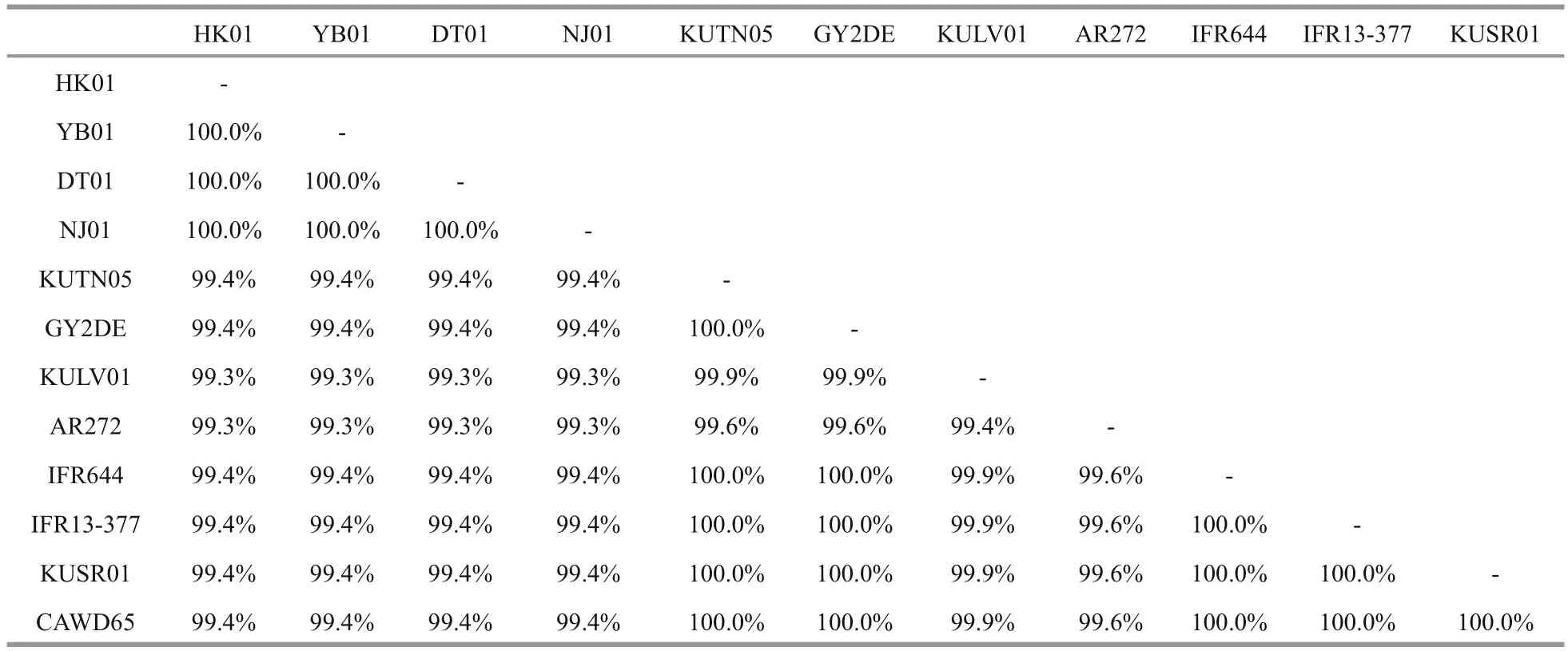
Table 3 Partial LSU sequence comparison among different Karenia longicanalis/ K. umbella
3.3 Molecular phylogeny
LSU rDNA sequences of the four strains DT01,YB01, NJ01 and HK01 (941 base pairs) ofK.longicanaliswere obtained and deposited in GenBank under accession numbers KY216192 to KY216194 and KY287670. The four strains of this study share identical LSU rDNA sequences with a similarity of 99.4% to the holotype ofKareniaumbella(strain KUTN05, AY263963) (Table 3). The ML and BI methods yielded similar phylogenetic trees, hence only the ML tree is shown (Fig.6). Our strains(boldface in Fig.6) grouped with other strains identified asK.umbellawith support values of 61/0.89(bootstrap values for ML analysis/posterior probabilities for BI analysis), with cultures from Australia, eastern North America, France and New Zealand clustering in a separate clade. All sequences clustered with the otherKareniaspecies with strong support (99/1).
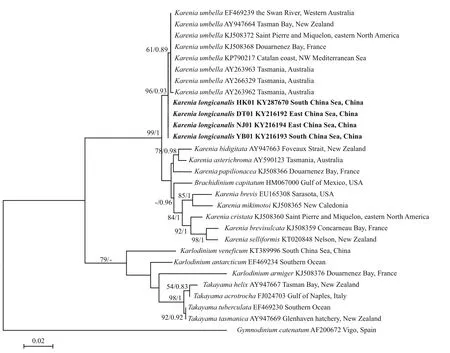
Fig.6 Phylogenetic tree (maximum-likelihood, ML) of Karenia longicanalis and its closely related species based on partial LSU sequences (578 bp in the final alignment)
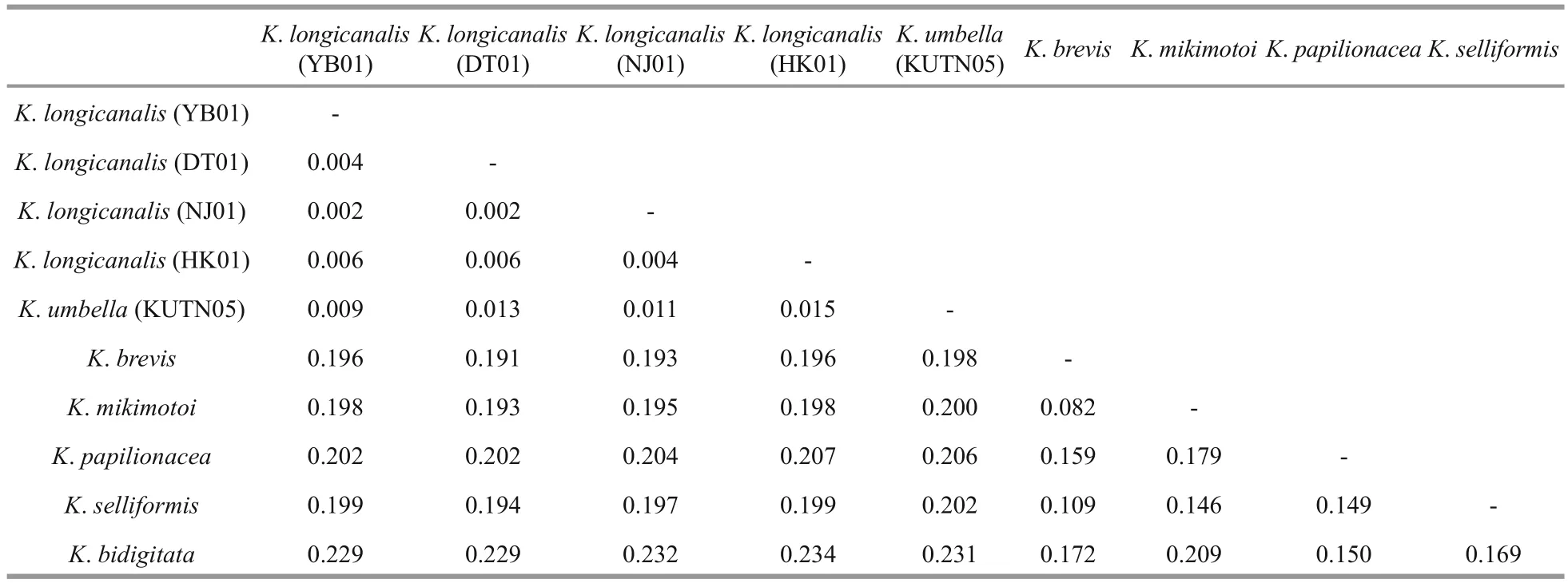
Table 4 Sequence divergence among different Karenia species based on ITS sequences (507 bp)
ITS sequences of the four strains (YB01, DT01,NJ01 and HK01) ofK.longicanaliswere obtained and deposited in GenBank under accession numbers MF781065 to MF781068. The divergence among different cultures ofK.longicanalis/K.umbellaranged from 0.002 to 0.015, while the congeneric interspecific divergence ranged from 0.082 to 0.234(Table 4). The ML and BI analysis based on ITS sequences generated similar phylogenetic tree (Fig.7).Sequences of our cultures clustered with the holotype ofK.umbellawith strong support (98/0.99).
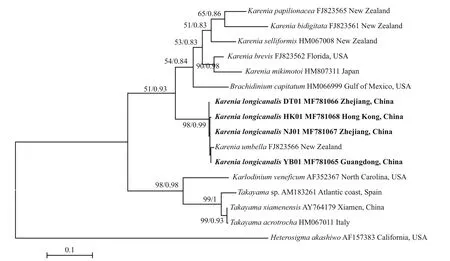
Fig.7 Phylogenetic tree (maximum-likelihood, ML) of Karenia longicanalis and its closely related species based on ITS sequences
4 DISCUSSION
Kareniaumbellawas recognized as a separate species based on cell morphology, pigment composition and molecular genetics (de Salas et al.,2004a). A bloom ofK.umbelladrew attention to the species when it caused mortality of caged rainbow trout in Murdunna, Australia in December 1989. A subsequent, more serious mortality event led to death of 100 000 Atlantic salmon in Tasmania, Australia in May 2003 (de Salas et al., 2004a). Till now,K.umbellahas been reported from Australia (de Salas et al., 2004a), New Zealand coastal waters (Chang et al.,2012), France and eastern North America (Nézan et al., 2014) and Japan (Omura et al., 2012).
Kareniaumbellais a relatively large species of the genusKarenia. The most typical features of this species are the size, the conical epicone, the asymmetrical hypocone and the eight radial furrows on the epicone. Morphological characteristics ofK.umbellaare, however, very similar to those ofK.longicanalis, a bloom-forming species described from Victoria Harbour, Hong Kong (Yang et al.,2001).K.umbellawas described to differ fromK.longicanalisin size, the shape of epicone and hypocone, the sulcal intrusion, the presence of epiconal furrows, and the shape and number of chloroplasts (Table 5) (Yang et al., 2001; de Salas et al., 2004a).
According to de Salas et al. (2004a),K.umbellais clearly larger thanK.longicanalis. The cell length and width ofK.umbellawere given by de Salas et al.(2004a) as 29–42 μm (average 35.85) and 21–32 μm(average 26.61), respectively, while those ofK.longicanaliswere 17.5–35 μm (average 25.9) and 10–22.5 μm (average 21.1) (Yang et al., 2001). In our study, the DT01 and YB01 strains were similar in size toK.umbellain de Salas et al. (2004a), but the NJ01 strain was similar toK.longicanalisin Yang et al.(2001) that measured field cells. There are no significant differences in size between our strains ofK.longicanalisandK.umbella. It should be noted that the size of the cell may vary with the culturing conditions.
Yang et al. (2001) described a critical feature ofK.longicanalisto be its extremely long apical groove which extended down over the upper level of the cingulum. In this study, the apical groove was equally long and reached the same level as inK.longicanalis.De Salas et al. (2004a) mentioned that the length of the apical groove on the dorsal side of the two species is different, the ratio inK.longicanalisbeing 2/3 down the epicone, whereasK.umbellait was 1/2.However, the length of apical groove may be affected by fixation during the preparation procedure of SEM,and our study revealed that the ratio of our material ranged from 1/2 to 2/3.
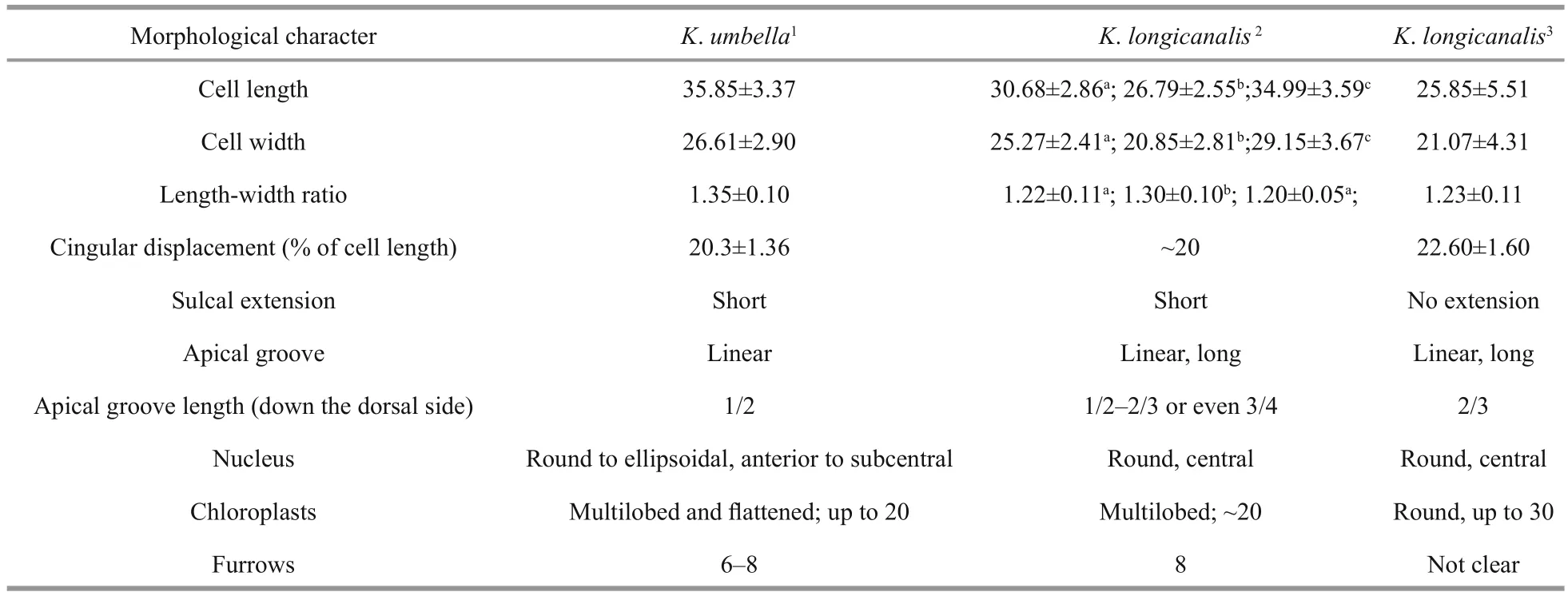
Table 5 A morphological comparison between Karenia umbella and Karenia longicanalis
The sulcal extension is also considered to be an important distinguishing feature, asK.umbellawas described with a short and finger-like sulcal intrusion onto the epicone, angling about 45° to the sulcus,while no sulcal intrusion was described inK.longicanalis. The sulcal extension ofK.umbellacould easily be observed under the LM by de Salas et al. (2004a), but in their SEM, notably Fig.I, there is no sulcal extension. A sulcal extension was not observed inK.longicanalis, neither in LM nor in SEM. In our study, we found that in cells in healthy state, the sulcal intrusion could be easily observed under LM. In the SEM, when our material was fixed in OsO4, cells were well preserved and the sulcal intrusion clearly visible. However, in cells fixed in glutaraldehyde or Lugol’s solution, the cells were more easily deformed; the sulcal intrusion was short or even missing (e.g. Fig.4a–c). Yang et al. (2001)examined cells by SEM that were preserved using the same Lugol’s solution concentration as in our study.We surmise that the Lugol’s solution fixative and the SEM preparation may be the reasons for the absence of a sulcal extension inK.longicanalis. In fact, Yang et al. (2001) mentioned that “the very delicate sulcal projection could have been lost inKarenialongicanalisduring the preparation for scanning electron microscopy”.
De Salas et al. (2004a) describedK.umbellaas having approximately 20 peripherally located,irregular and multi-lobed chloroplasts, whereasK.longicanalishad about 30 globular chloroplasts.We observed that in our material, chloroplasts were irregular and multi-lobed, while chloroplasts in unhealthy state, such as barely moving cells, the chloroplasts became globular. Chloroplasts ofK.umbellapresented by Omura et al. (2012) are globular and very similar to those described inK.longicanalis. In someKareniaspecies, the chloroplast becomes circular in shape when fixed (Botes et al.,2003); we also found that chloroplasts of our material fixed in Lugol’s solution became somewhat rounded.Therefore, the chloroplast is not a reliable character to differentiate the two species.
Kareniaumbellawas characterized by eight radial furrows on the epicone when seen in the SEM, which gave rise to the species name (de Salas et al., 2004a).However, the furrows are very easily lost during the preparation of SEM. We have tested samples fixed in different fixatives for SEM, such as Lugol’s solution,glutaraldehyde and OsO4, and found that most cells fixed in glutaraldehyde and Lugol’s solution have lost the furrows during the preparation, only cells fixed in OsO4solution retained the furrows. Furrows were not observed inK.longicanalis; whether this is due to the cells being fixed in Lugol’s solution is not known but it is likely. In another species of the Kareniaceae,Karlodiniumgentienii, Nézan et al. (2014) observed a somewhat similar phenomenon. In some fixation cocktails numerous radiating striae were clearly visible on the epicone while in others no such striae could be observed. Another puzzling feature in whichK.umbellais thought to differ fromK.longicanalisis the presence of several deep pores on the left ventral hypocone or epicone, or both (de Salas et al., 2004a).The number of pores was variable and they are not easily preserved in the SEM. We observed these pores only in the hypocone, showing a variable number.The role of the pores is uncertain as well as its use as a diagnostic character.
No information on DNA sequences ofK.longicanalisis available in GenBank and the species was not obtained in culture when described from Victoria Harbour, Hong Kong by Yang et al.(2001). A comparison of the partial LSU rDNA of a strain identified asK.longicanalis(strain HK2) from Hong Kong andK.umbellafrom New Zealand (strain CAWD65 and GenBank No. AY947664) performed by Gu (2007) revealed that the two species have a high similarity of 99.43% and only differed in 4 out of the total 713 bases sequenced, suggesting the two species to be conspecific. In this study, the similarity among different cultures ofK.longicanalis/K.umbellaranged from 99.3% to 100.0% (based on partial LSU sequences, 708 bp). The four Chinese strains share identical LSU rDNA sequences with a similarity of 99.4% to the holotype ofK.umbella(strain KUTN05,AY263963) and as well as to the strain CAWD65 used in Gu (2007), and they are also different in 4 bases. Therefore, the morphological comparison and the phylogenetic data confirm thatK.longicanalisandK.umbellaare conspecific.
5 CONCLUSION
Morphological and molecular of the data reveal that there is no reliable difference betweenK.longicanalisandK.umbella. We conclude that these species are synonyms with priority forK.longicanalis.
6 DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
7 ACKNOWLEDGEMENT
We sincerely thank Dr. GUAN Wanchun of Wenzhou Medical University for assistance in the field sample collection. Special appreciation is expressed to Dr. LUO Zhaohe of the Third Institute of Oceanography, the State Oceanic Administration for helpful discussions regarding the SEM preparation.We would also like to thank XIA Xiaofei of Beijing Museum of Natural History for his support during SEM photography.

Table S1 Information of LSU rDNA sequences used in this study for phylogenetic analysis
Table S1 Continued
Table S2 Information of ITS sequences used in this study for phylogenetic analysis
 Journal of Oceanology and Limnology2018年6期
Journal of Oceanology and Limnology2018年6期
- Journal of Oceanology and Limnology的其它文章
- Neuroanatomy and morphological diversity of brain cells from adult crayfish Cherax quadricarinatus*
- On the influence of season and salinity on the phenology of invertebrates in Australian saline lakes, with special reference to those of the Paroo in the semiarid inland
- Man-made plutonium radioisotopes in the salt lakes of the Crimean peninsula
- Cladophora mats in a Crimean hypersaline lake: structure,dynamics, and inhabiting animals
- Antimony speciation at the sediment-water interface of the Poyang Lake: response to seasonal variation*
- Seasonal variations of phosphorus species in the Tuohe River,China. Part I. Sediments*
