Pf- D mrt4, a potential factor in sexual development in the pearl oyster Pinctada f ucata*
WANG Qi (王琦) , SHI Yu (石禹) HE Maoxian (何毛贤)
1 CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
2 Taiyuan University of Technology, Taiyuan 030024, China
Abstract The mechanisms of sex determination and sex differentiation in the pearl oyster Pinctada fucata are currently poorly understood. We therefore investigated the roles of orthologs of the Dmrt gene family, key players in male gonad differentiation in mammals, in P. fucata sex differentiation and sexual development.Pf- Dmrt4 exhibits features typical of the D mrt family, and displays significant homologies to the DMRT4 cluster. Pf- Dmrt4 mRNA expression in the gonads during a gametogenic cycle, measured by quantitative polymerase chain reaction, was maximal in mature individuals. Pf- Dmrt4 expression, demonstrated by in situ hybridization, was localized in the spermatozoa, spermatids, oocytes and vitellogenic oocytes.Knockdown of Pf- Dmrt4 with double-stranded RNA resulted in decreased mRNA expression levels. And Pf-Dmrt4-dsRNA-injected groups showed spawning-stage male gonads, with ruptured follicles and released spermatozoa. Our results enhance the understanding of sex determination and differentiation in P. fucata and suggest that Pf- Dmrt4 could be involved in male gonadal development, and maintenance of male gonadal function.
Keyword: Pinctada fucata; Pf- Dmrt4; gene expression; sexual development
1 INTRODUCTION
Sexual reproduction is a remarkable phenomenon in biology and the primary source of genetic variation required for evolutionary survival. It can be found in almost all types of multicellular animals. The mechanisms of sex determination and differentiation vary among phyla, possibly as a result of long-term evolution of the genes involved in the sex determination pathways (Marín and Baker, 1998).Bivalves belong to the second largest phylum and display highly diverse sexual phenotypes. The mollusc pearl oysterPinctadafucataexhibits complex modes of sexual reproduction that consists of protandric dioecy, occasional sex change and hermaphroditism. Environmental factors such as population density, water temperature, and sex ratios play important roles in sex determination and differentiation in bivalves (Yu et al., 2007, 2011),though the function of genetic factors remains unclear.
TheDrosophiladoublesex(Burtis and Baker,1989) andCaenorhabditiselegansmab-3(Shen and Hodgkin, 1988) genes are widely conserved and have been implicated in sex determination (Raymond et al., 1999). TheDmrt(double-sex and mab-3-related transcription factor) gene family encodes proteins with conserved DM domains (a common zinc fingerlike DNA-binding motif) and is represented by multiple members in both vertebrates and invertebrates(Volff et al., 2003). To date, eight members of the family have been reported in humans (Ottolenghi et al., 2002) and seven have been found in mice (Kim et al., 2003). DM-domain family members have been shown to regulate sexual development in vertebrates and invertebrates, and the function of theDmrtgene family in sex regulation has been highly conserved during evolution (Hodgkin, 2002).
Apart fromDmrt1, DM-domain family members are expressed during development in tissues other than the gonads, suggesting that these proteins may control a broader range of developmental processes(Hong et al., 2007). SomeDmrt4andDmrt5orthologs play key roles in sexual development in different animals such as zebrafish (Guo et al., 2004; Winkler et al., 2004), blue tilapia (Cao et al., 2010),Chlamys farreri(Feng et al., 2010) andCrassostrea.gigas(Naimi et al., 2009).
As a highly conserved gene family,Dmrtgenes have been cloned and studied in various vertebrates(Ottolenghi et al., 2002; Aoyama et al., 2003) and several invertebrates, including fruit flies (Crémazy et al., 2001), nematodes (Volff et al., 2003), sea squirts(Leveugle et al., 2004) and freshwater prawns (Peng et al., 2005). In molluscs, orthologs of the DM-domain transcription factors have been isolated from the oysterC.gigas(CgDMl) (Naimi et al., 2009), the scallopCh.farreri(Cfdmrt4-like) (Feng et al., 2010),the abaloneHaliotisasinina(HADMRT1-like)(Klinbunga et al., 2009), the pearl oysterPinctada martensii(pmDmrt2andpmDmrt5) (Yu et al., 2009,2011), and the scallopCh.nobilis(dmrt5) (Shi et al.,2014). Gene expression analysis of some of these orthologs suggests the possible involvement of these genes in gonad differentiation and development(Naimi et al., 2009; Feng et al., 2010). Sex determination and differentiation in molluscs appear to be complex because of the diverse range of species and underlying molecular mechanisms involved.
Recently, the genomics of some economically marine bivalves are studied to provide better understanding of the molecular mechanisms underlying their different reproductive strategies such as the black-lip pearl oysterP.margaritifera(Teaniniuraitemoana et al.,2014), the Pacific oysterC.gigas(Zhang et al., 2014) and the pearl oysterP.fucata(Takeuchi et al., 2012; Du et al., 2017). The available transcriptome is a powerful and novel resource to study sexual pathway in marine mollusks.
It is an essential step to identify and characterize sex-specific genes to explore the molecular pathways of sexual development in the pearl oysterP.fucata.This study therefore investigated the genetic mechanisms responsible for sex differentiation and gonad development ofPf-Dmrt4inP.fucata, which is an economically important species for pearl production in China. The full-length cDNA sequence of Pf-Dmrt4 during gonad differentiation was characterized, and the spatio-temporal expression patters ofPf-Dmrt4were studied during gonad differentiation and in adult tissue using real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The cellular localization and the sex-steroid –induced expression of the gene were also investigated.
2 MATERIAL AND METHOD
2.1 Experimental animals and tissue sampling
Pearl oystersP.fucatawere obtained from Daya Bay in Shenzhen (Guangdong Province, China),cultivated in floating net cages in the sea under natural conditions. All specimens were collected between May, 2012 and August, 2013. Testes and ovaries were excised, frozen immediately in liquid nitrogen and maintained at -80°C until RNA extraction forPf-Dmrt4cloning. Ovary, testis, heart, digestive gland,adductor muscle, mantle, and gill tissues were also dissected and stored in the same manner for tissuedistribution analysis. Gonad tissues from different stages were dissected, fixed in Bouin’s solution at 4°C overnight, and submitted to histological observation to determine the gonadal stages. Gonad samples were fixed with 4% paraformaldehyde dissolved in phosphate-buffered saline (PBS) overnight at 4°C and then stored in methanol at -20°C for in- situ hybridization. Gametes were obtained and passed through a 100-μm screen to remove tissue debris. The eggs were mixed with sperms at 25–26°C in filtered seawater which contained 0.005%–0.006% (v/v)ammonia. After fertilization, embryos and different stages of larvae were then prepared for RNA extraction.
2.2 Cloning and sequencing of Pf- Dmrt4
Total RNA was extracted from testes using an E.Z.N.A. Mollusc RNA Kit according to the manufacturer’s protocol (Omega, Japan). The quantity and quality of RNA were assayed by UV spectrophotometry (OD260/OD280). A ReverTra Ace-First-strand cDNA Synthesis Kit (Toyobo, Japan) was used to synthesize first-strand cDNA from 1 μg total RNA. All primers forPf-Dmrt4used in the present study are listed in Table 1. PCR primers were designed using Primer Premier 5.00 (Premier Biosoft International, Palo Alto, CA, USA). All primer pairs were designed to eliminate false-positive bands of potential genomic DNA contamination. A fragment obtained from the transcriptome ofP.fucatawas amplified using a pair of specific primers df 1 and dr1.BD SMART RACE cDNA Amplification Kit(Clontech, USA) was used to synthesize SMART cDNA. The resulting 1 384-bp fragment was used to generate full-length cDNA by 5′- and 3′ rapid amplification of cDNA ends. The cycling conditions for all PCR were: 3 min at 94°C for denaturation,35–40 cycles at 94°C for 30 s, 55–60°C for 30 s and 72°C for 1–2 min, followed by a further elongation at 72°C for 10 min. The PCR products were purified with The E.Z.N.A. Gel Extraction Kit (Omega BioTek, USA), subcloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA) and sequenced(Applied Biosystems, USA).
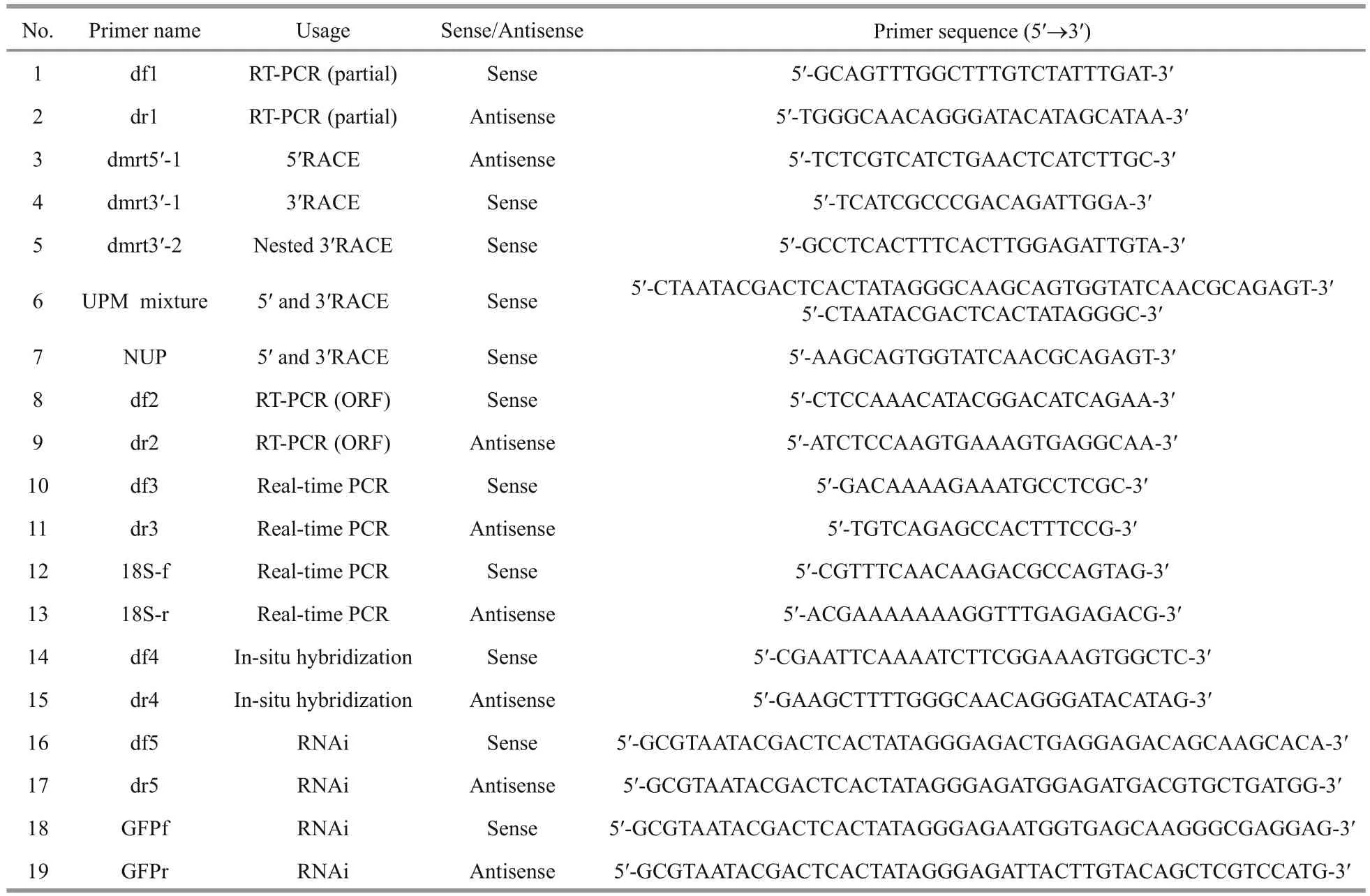
Table 1 Primers used for Pf- Dmrt4 cloning and functional analysis
2.3 Tissue distributions of Pf- Dmrt4 in P. fucata
Tissue expression patterns ofPf-Dmrt4mRNA were analyzed by real-time qPCR. Expression analysis was performed in ovary, testis, gill, mantle,heart, digestive gland, and adductor muscle tissues collected from three pearl oysters and stored at -80°C.Reactions were performed in triplicate. Real-time qPCR were carried out using a Roche LightCycler 480 real-time PCR system, with SYBR green master mix (Toyobo, Japan) following the manufacturer’s instructions. All the qPCR primers are presented in Table 1. Real-time PCR was performed as follows:94°C for 1 min, and 40 cycles of 94°C for 15 s, 15 s at 55°C and 72°C for 1 min.
2.4 Expression patterns of Pf- Dmrt4 in embryonic and larval P. fucata
The developmental expression patterns ofPf-Dmrt4were analyzed in five developmental stages,which were collected and stored at -80°C: blastulastage embryo, trochophore, D-shaped larva, umbo larva and metamorphosis stage. Total RNA extraction and real-time qPCR were the same as described above and three repetitions were analyzed.
2.5 Expression patterns of Pf- Dmrt4 in P. fucata gonads during gametogenesis
Gonadal sections were fixed with Bouin’s fixative ,6 μm slices were prepared, and processed with conventional histological analysis. The sections were stained with hematoxylin-eosin and examined microscopically to classify the natural gonadal development. The gametogenic expression patterns ofPf-Dmrt4were analysed by real-time PCR in cDNAs from four stages of ovary and testis (growing stage, mature stage, spawning stage and resting stage), synthesized as described above. Five replicates of each sample were analyzed.
2.6 Expression of Pf- Dmrt4 mRNA following methyltestosterone injection
The effect of methyltestosterone (MT) on the mRNA expression levels ofPf-Dmrt4were investigated by injecting male oysters with MT(purchased from Guangzhou Lifescience Biotechnology Co. Ltd., Guangzhou, China) at the gonadal mature stage. MT was dissolved in dimethyl sulfoxide (DMSO) (Sigma, St. Louis, MO) and added into modified Herbst’s artificial seawater (ASW) to a concentration of 1 μg/μL. A 100-μL aliquot of suspension was injected into the adductor muscle of male oysters. The animals in Negative control group were injected with 100 μL of ASW containing 1%DMSO. Eight oysters were randomly collected before injection (blank control). A further eight oysters were collected every 6 h after injection for 3 times,respectively, and the gonads were dissected and fixed for RNA extraction and real-time PCR.
2.7 In situ hybridization
The expression ofPf-Dmrt4in gonads were localized using the gene-specific primers designed for amplification (Table 1). Digoxigenin -labeled RNA probes were prepared from a 482-bp fragment of thePf-Dmrt4cDNA that had been amplified and subcloned into pGEM-T Easy vectors (Promega) as a template. The probes were labeled with a DIG RNA labelling kit (Roche Diagnostics, Switzerland).Transcriptions were performed using SP6 (sense) or T7 (antisense) RNA polymerase. Paraformaldehydefixed gonad samples were embedded in paraffin,sectioned (10 μm), de-paraffinized, and then rehydrated. In situ hybridization was performed as described by Miyashita et al. (2008), with some modifications. Gonad sections were hybridized with DIG-labeled RNA probes, and hybridized signals were visualized with NBT/BCIP system and alkaline phosphatase-conjugated anti-DIG antibody using a DIG Nucleic Acid Detection Kit (Roche Diagnostics)following the manufacturer’s protocol.
2.8 RNA interference
RNA interference was performed as described by Suzuki et al. (2009), with some modifications. The primers used to generatePf-Dmrt4and green fluorescent protein (GFP) double-stranded (ds) RNAs are shown in Table 1. The T7 promoter sequence is underlined. RiboMAXTM Large Scale RNA Production System (T7) kit (Promega, USA) was uesd to synthesize and purify the dsRNA. RNase-free DNase I (TaKaRa, Japan) was used to digest template DNA. Synthesized dsRNA was diluted to 10, 20 and 40 μg/100 μL with PBS, and 100 μL of each solution was injected into the adductor muscles of 2-year-old oysters with mature testes. PBS 100 μL or 20 μgGFPdsRNA in 100 μL PBS were used as blank and negative controls. Each treatment selected 7 oysters.Total RNA was extracted from the gonad of each individual 7 days after injection and used for firststrand cDNA synthesis. The expression levels were quantified by real-time qPCR, using18Sas a reference gene. ThePf-Dmrt4qPCR primers were designed to amplify the region that did not overlap the region targeted by dsRNA. Gonads were dissected from the injected-oyster groups 7 days after injection, fixed in Bouin’s solution at 4 °C overnight, and submitted to histological observation at the light-microscope level.
2.9 Statistical analysis
Homology analysis of the deduced amino acid (aa)sequence was conducted using the public NCBIBLAST database. Motif analysis was performed using InterPro Scan on the ExPASy Proteomics Server. ClustalX (1.81) was used for multiple sequence alignments of amio acids. A phylogenetic tree was constructed by the neighbor-joining method using MEGA.6. Three-dimensional model of Pf-DMRT4 were predicted by SWISS Model on http://swissmodel.expasy.org/. All reference sequences were downloaded from GenBank and public genome resources. Quantitative data were shown as mean±S.E.M. Significant differences were estimated by one-way ANOVA followed by Duncan’s multiple range tests.
3 RESULT
3.1 Isolation and characterization of Pf- D mrt4
A full-length cDNA encodingPf-Dmrt4was isolated fromP.fucata(GenBank accession number:KM272582). The cDNA sequence ofPf-Dmrt4was 1 711 bp, containing an open reading frame of 1 104 bp, 5′ untranlated region (UTR) of 199 bp and 3′UTR of 408 bp with a 30-bp polyA tail. The deduced aa sequence was 367 aa and contained a DM domain consensus sequence (from 23–81 aa) with conserved cysteines and histidines characteristic of the DMRT protein family, and a DMA domain (from 207–242 aa)(Fig.1).

Fig.1 Nucleotide and deduced amino acid sequences of Pf- Dmrt4
The amino acid sequence of the conserved DM domain ofP.fucataPf-DMRT4 was aligned with known sequences in various members of this family in different species (Fig.2a). The highest identity rate was 100% withP.martensiiDMRT5, followed byCh.farreriDMRT4-like,C.gigasDMI andCh.nobilisDMRT5 (98%). Sequence comparison revealed a conserved zinc module consisting of intertwined CCHC and HCCC Zn2+-binding sites, and a consensus sequence of five aa (KGHKR) of the putative nuclear localization signal (NLS). There was a conserved DMA domain and a short conserved domain of seven aa (KSAFSPI) near the C-terminus in DMRT4 and DMRT5, but not in DMRT-like types (Fig.2b). The DMA domain ofP.fucataDMRT4 displayed highest homologies of 74% with Cg-DMI, 68% withCh.farreriDMRT4-like, 66% withCh.nobilisDMRT5 and 65% withOryziaslatipesDMRT5. The seven-aa conserved domain shared the highest levels of aa identity withCh.farreriDMRT4-like andCh.nobilisDMRT5 (100%), Cg-DMI andH.sapiensDMRT4(85.71%) andH.sapiensDMRT5 (71.43%).

Fig.2 Alignment of amino acid sequences of the DM domain of P. fucata DMRT4 with other DMRTs of different species (a)and alignment of amino acid sequences of the DMA domain and the short conserved motif from different species (b)
The phylogenetic tree generated using the complete protein genes of DMRT from various species showed thatP.fucataDMRT4 was most closely related toP.martensiiDMRT5, then clustered withCh.farreriDMRT4-like,C.gigasDMI andCh.nobilisDMRT5(Fig.3a).P.fucataDMRT4 was grouped with the mollusc DMRT4 and DMRT5 clusters, but more closely with DMRT4, implying thatP.fucataDMRT4 may be a member of the DMRT4 cluster. Two transcripts encoding orthologs of the DM domain transcription factor were identified inP.margaritifera,PinMarg DMRT1 and PinMarg DMRT2 were clustered with OryLat DMRT1 and PinMar DMRT2,then grouped with DMRT1 isoform X1 and X2 which were found in the genomic database inC.gigas.
A model of the structure of the Pf-DMRT4 was predicted by SWISS Model and the yellow indicated zinc-finger-like with DNA binding modules of DM domain (Fig.3b).

Fig.3 The phylogenetic tree of the DMRT proteins was constructed by MEGA.6 using the neighbor-joining method (a) and structure model of Pf-DMRT4 protein (b)

Fig.4 Expressions of Pf- Dmrt4 mRNA in tissues of P. fucata (a); expression pattern of Pf- Dmrt4 mRNA in embryonic and larval stages of P. fucata (b); Pf- Dmrt4 mRNA temporal expression in adult gonadic areas of P. fucata along a gametogenetic cycle (c); levels of mRNA expression for Pf- Dmrt4 mRNA in the gonad of P. fucata after MT injection(d); expression levels of Pf- Dmrt4 knocked down by RNAi (e)
3.2 Pf- Dmrt4 expression pattern in tissues
Pf-Dmrt4mRNA expression levels were detected in all tested tissues by real time qRT-PCR. The expression profiles using df3, dr3 and Sfi , Sr1 primers suggested that mRNA expression reflectedPf-Dmrt4transcript levels.Pf-Dmrt4mRNA levels were significantly higher in gills and testis, moderate in digestive gland, and low in mantle, adductor muscle and heart tissues. In female tissues,Pf-Dmrt4mRNA levels were higher in the mantle and ovary compared with other tissues.Pf-Dmrt4expression levels were higher in testis than in ovaries (Fig.4a).
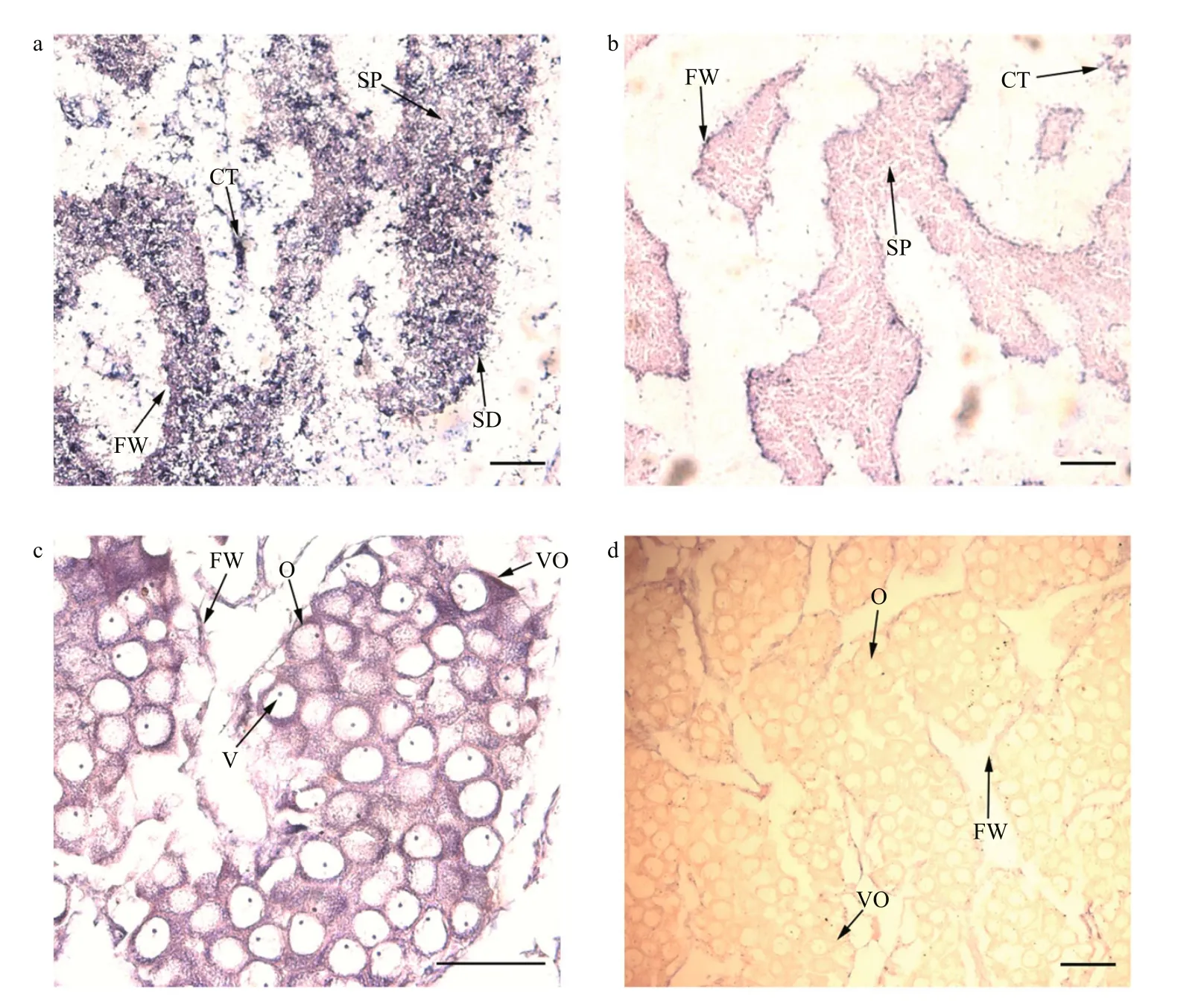
Fig.5 Pf- Dmrt4 mRNA cellular expression patterns by in-situ hybridization in the gonadic area during the mature stage
3.3 Pf- Dmrt4 expression patterns in different developmental stages of P. fucata
Pf-Dmrt4mRNAs were expressed in all embryonic and larval developmental stages examined (Fig.4b).Pf-Dmrt4expression was highest in D-shaped larvae.
3.4 Temporal expression patterns of Pf- Dmrt4 during the adult gametogenic cycle
The gametogenic cycle can be divided into four stages: growing stage (active spermatogenesis and growing oocytes), mature stage (mature germ cells and almost no follicular internal), spawning stage(ruptured follicular, spawning mature sperms and oocytes) and resting stage (irregular shape of follicular, contains few spermatogonia and oogonia,degenerate oocytes and sperms can be seen occasionally).Pf-Dmrt4mRNA expression levels in gonad were detected by real-time qPCR at all adult gametogenic stages (Fig.4c).Pf-Dmrt4mRNA levels were significantly higher in the mature stage compared with the other three stages in the male gametogenic cycle. However, there were no significant differences inPf-Dmrt4expression among the different stages of the female gametogenic cycle.
3.5 Effects of MT injection on Pf- Dmrt4 expression
Pf-Dmrt4mRNA expression in the gonads during a gametogenic cycle, measured by quantitative polymerase chain reaction, was maximal in mature male testis. Therefore, mature male testis were used in the MT injection experiment.
Pf-Dmrt4mRNA levels in the testis were significantly increased 6-h post-injection with 1 μg/μL of MT (Fig.4d), being almost 3- fold higher than in control groups, respectively.
3.6 Localization of Pf- Dmrt4 transcripts in the gonads of P. fucata
We determined the cellular localizations ofPf-Dmrt4transcripts in matureP.fucatabyinsituhybridization.Pf-Dmrt4mRNA in the male gonads was detected in the cytoplasm of spermatozoa and to a lesser extent in spermatids (Fig.5a). In the female gonads (Fig.5c), cytoplasmic mRNA staining was observed in oocytes and vitellogenic oocytes.Negative controls with the sense riboprobe sometimes produced a faint, insignificant signal (Fig.5b, d).

Fig.6 Light microscope images of the testis of the pearl oysters injected with PBS (a), 20 μg of Pf- Dmrt4 (b) dsRNA in the magnification of 100, respectively
3.7 RNA interference knockdown of Pf- Dmrt4
Pf-Dmrt4expression levels were decreased byPf-Dmrt4dsRNA (10, 20, and 40 μg) respectively(Fig.4e). Expression levels in the groups injected with 20 μgPf-Dmrt4dsRNA were suppressed to approximately 70% and 50% of those in the PBS- and GFP-dsRNA-injected groups, respectively.
Male gonads of pearl oysters in each injection group were observed under an optical microscope.The gonad structure was normal (mature stage) in the PBS-injected groups (Fig.6a), whilePf-Dmrt4-dsRNA-injected groups showed spawning-stage male gonads (Fig.6b), with ruptured follicles and released spermatozoa.
4 DISCUSSION
4.1 Structure and molecular characteristics of Pf-Dmrt4
TheDmrt-family genePf-Dmrt4from the gonads ofP.fucatawas cloned. The deduced aa sequence of Pf-Dmrt4 is serine- and proline-rich, includes a DM domain characteristic of the DMRT protein family, a conserved DMA domain near the C-terminus, and a short seven-aa motif (KSAFSPI). The DM domain contains the putative NLS (KGHKR) located in the zinc module, consisting of intertwined CCHC and HCCC Zn2+-binding sites. The NLS is characterized as a transcription regulator of the DMRT family and plays a part in DNA-binding and nuclear import(Zhang et al., 2001; Ying et al., 2007). The DMA domain is conserved in the branch of DMRT3,DMRT4 and DMRT5 (Guo et al., 2004). Wen et al.(2009) assumed that the DMA domain may act as a motif to bind other transcriptional co-factors and can probably be essential for the role of DMRT4 to regulate other genes in transcriptional level.
Yu et al. (2009) previously cloned PinMarDmrt5 fromP.martensii, which is largely identical toPf-Dmrt4. However, the deduced PinMarDmrt5 product of 206 aa lacks the DMA domain and the short conserved motif. Sequence alignment of the two genes revealed that PinMarDmrt5 lacked four nucleotides causing a frame shift. The subsequent nucleotide sequence resulted in a stop codon, thus explaining the short product with no DMA domain or conserved motif.P.fucataDMRT4 displayed higher homologies with human DMRT4 and DMRT5 than with human DMRT1–3.
The availability of theP.fucatagenome (Takeuchi et al., 2012; Du et al., 2017) provide an opportunity for a comprehensive study of sex-determining pathways in this species. In the genome of theP.fucata, a dmrtA2-like gene was investigated. While pacific oyster genome encodes three DM domain containing genes:Cg19568,Cg01830, andCg15952(namedCgDMl) (Zhang et al., 2014). They found that high expression in testis supports a possible role forCg19568(namedCgDsx) in determining or promoting male-specific development. It contains a DM domain showing closest homology (45%, E-value=9e-13) toDsxisoform A found inD.melanogaster.Pmargdmrt2sequence was seen to share the highest amino acid identity withP.martensiidmrt2and zebrafishDmrt2(100% and 95%, respectively), andpmarg-dmrtwithP.martensiidmrt2and mouseDmrt4(59%and 58%, respectively). The other two DM domain genes both show the highest homology to DmrtA2 from many species. Phylogenetic analysis showed thatPf-Dmrt4was grouped with DMRT4 clusters and more closely with human DMRT4.Pf-Dmrt4isa DM domain gene and a close relative of CgDMl. Volff et al. (2003) proposed a simpler nomenclature (Dmrt1–8) based on human genes, with no suggestions of structural or phylogenetic relationships between different genes. We therefore named it asPf-Dmrt4.
4.2 Potential function of Pf- Dmrt4
It was reported that ancestral DM proteins possessed a DMA domain (Miller et al., 2003; Volff et al., 2003), and the presence of a DMA domain inPf-DMRT4 reinforces the hypothesis.Pf-DMRT4 as an ancestral DM factor, could be engaged in a number of biological processes, including functions associated with the ancestral structure, whereas evolution has led to diverse DM factors.
Pf-Dmrt4was expressed in all the tested adult tissues, which is similar toDMlinC.gigas(Naimi et al., 2009),Cf-dmrt4-likeinCh.farreri(Feng et al.,2010), and related genes in other species with different expression patterns (Kim et al., 2003; Guo et al.,2004). Differences in DMRT4 expression among species might reflect variations in gene function. InTakifugurubripes,Dmrt4is expressed in the spleen,suggesting an involvement in the immune system(Yamaguchi et al., 2006). Additionally,Pf-Dmrt4mRNA expression levels were highest in male gills,as fordmrt5inCh.nobilis(Shi et al., 2014). While,dmrt5was expressed highest in gills but no sexual difference has been detected inCh.farreri(Feng et al., 2010) and inC.gigas(Naimi et al., 2009). These findings suggested thatPf-Dmrt4is involved in the respiratory system.
Pf-Dmrt4mRNA was also expressed widely in embryonic and larval stages. Its detection at an early stage resembled the situations inCh.farreri(Feng et al., 2010),Ch.nobilis(Shi et al., 2014), and some other vertebrates such as medaka (Winkler et al.,2004).Cf-dmrt4-likeexpression was ubiquitous during embryonic development and peaked at the 8–16-cell stage (Feng et al., 2010), whileCh.nobilis dmrt5expression peaked at the blastula stage (Shi et al., 2014).Pf-Dmrt4mRNA expression peaked at the D-shaped larva stage, and this stage is a crucial period to build the larva structure, indicating thatPf-Dmrt4may play a role in early embryonic development,especially in ontogenesis of larva.
Pf-Dmrt4may also be involved in gonadal development in the pearl oyster. It was expressed in both sexes, but at higher levels in the testis than in the ovary, as reported forDmrt4inCh.farreri(Feng et al., 2010),DMlinC.gigas(Naimi et al., 2009),medaka (Winkler et al., 2004), and the Japanese pufferfish (Yamaguchi et al., 2006).Pf-Dmrt4expression levels increased significantly during the mature stage in the testis, but there were no significant differences in mRNA expression in the female gonad,consistent with an increase inCg-DMlin males at the mature stage (Naimi et al., 2009). The number of spermatogonia increase in the mature stage (stage III), as a result not only of their proliferation but also of an increase in gonad volume from 5%–40% of the visceral mass in less-mature animals to 60% at this stage (Fabioux et al., 2004). This indicates thatPf-Dmrt4may be involved in regulating male gonadal development inP.fucata.
In situ hybridization of the testis revealed thatPf-Dmrt4was expressed in the cytoplasm of spermatozoa and spermatids, though it was impossible to differentiate between germ cell and somatic cell limits at this microscopic level. InC.gigas,Cg-DMlwas observed in the cytoplasm of spermatogonia and/or the surrounding somatic cells (Naimi et al., 2009), in agreement with the expression ofDmrt5in testicular germ cells in zebrafish (Guo et al., 2004). Different expression patterns exist in different species(Raymond et al., 2000; Winkler et al., 2004).Variations inPf-Dmrt4localization suggest that it may play divergent roles in the gonad, including involvement in the proliferation of spermatozoa.Knockdown ofPf-Dmrt4by RNA interference caused morphological changes in the male gonad, indicating thatPf-Dmrt4might be involved in maintaining the structure of the male gonad inP.fucata.
Sex steroids play an important role in bivalve reproduction and can promote gamete development.Estrogen injection can affect gametogenesis and gametogenesis-related metabolic pathways in invertebrates (Wang and Croll, 2003; Croll and Wang,2007). MT significantly upregulatedPf-Dmrt4expression levels in mature-stage male gonads.Though the effects of sex steroid hormones onDmrt4orDmrt5are largely unknown, Shi et al. (2014)showed thatDmrt5mRNA inCh.nobilismale gonads was significantly increased by injection with three different concentrations of MT. Similar findings were observed forOreochromisaureusDmrt4, implying thatDmrt4might be a direct downstream gene under the control of steroids (Cao et al., 2010), and thatPf-Dmrt4might play a similar role.
5 CONCLUSION
Pf-Dmrt4is involved in the sex-differentiation pathway in the pearl oysterP.fucata.Pf-Dmrt4is involved in a wide range of biological processes,including cell proliferation and/or regulating male gonadal development and maintaining the structure of the mature testis inP.fucata. More studies are needed to clarify the precise physiological functions ofPf-Dmrt4.
6 DATA AVALABILITY STATEMENT
Sequence data that support the findings of this study have been deposited in National Center for Biotechnology Information (GenBank) with the primary accession code KM272582.
Electronic supplementary material
Supplementary material (Supplementary Tables S1–S3, Figs.S1–S7, Texts S1–S3) is available in the online version of this article at https://doi.org/10.1007/s00343-019-7305-z.

Supplementary Material 1 GenBank accession numbers of the reference sequences used in the Fig.2

Supplementary Material 1 Continued

Supplementary Material 2 Abbreviations of the species used in Fig.3
 Journal of Oceanology and Limnology2018年6期
Journal of Oceanology and Limnology2018年6期
- Journal of Oceanology and Limnology的其它文章
- Neuroanatomy and morphological diversity of brain cells from adult crayfish Cherax quadricarinatus*
- On the influence of season and salinity on the phenology of invertebrates in Australian saline lakes, with special reference to those of the Paroo in the semiarid inland
- Man-made plutonium radioisotopes in the salt lakes of the Crimean peninsula
- Cladophora mats in a Crimean hypersaline lake: structure,dynamics, and inhabiting animals
- Antimony speciation at the sediment-water interface of the Poyang Lake: response to seasonal variation*
- Seasonal variations of phosphorus species in the Tuohe River,China. Part I. Sediments*
