Development and Validation of Microsatellite Markers Derived from the Genome DNA Sequence of Larimichthys crocea
XIE Fei-ang ,ZHAO Ruo-ping ,HE Qi ,MAO Jun-lai ,WANG Yi-fan ,JIANG Li-hua ,SENANAN Wansuk,CHEN Yong-jiu,,,
(1.School of Marine Science and Technology of Zhejiang Ocean University,Zhoushan 316022,China;2.State Key Laboratory of Genetic Resources and Evolution,Kunming Institute of Zoology,Chinese Academy of Sciences,Kunming 650203,China;3.National Research Center for Marine Facility Aquaculture Engineering Technology,Zhoushan 316022,China;4.Department of Aquatic Science,Faculty of Science,Burapha University,Bangsaen,Chonburi 20131,Thailand)
Abstract:The large yellow croaker (Larimichthys crocea) and its sibling species,small yellow croaker(L.polyactis) are both important commercial drum fish species (Order:Perciformes;Family:Sciaenidae) native to the China Seas.In this study,we developed a total of 499,638 microsatellite DNA loci from the genome sequence of L.crocea.We evaluated polymorphisms of 22 selected tetra-nucleotide microsatellite loci in L.crocea(n=21) and L.polyactis(n=3).In L.crocea,a total of 220 alleles were scored with an average of 10±5.58 alleles(NA) per locus;the number of effective allele (NE),observed heterozygosity (HO),expected heterozygosity (HE)and polymorphic information content (PIC) averaged over loci are 5.40±3.68,0.64±0.25,0.71±0.22 and 0.67±0.22,respectively.Cross amplification in L.polyactis yielded 66 alleles in total (NA=3±1.51),and NE,HO,HE and PIC averaged over loci are 2.65±1.40,0.52±0.38,0.60±0.34 and 0.45±0.26,respectively.These microsatellite DNA markers could cross-amply and thus benefit advanced studies in population genetics and stock assessment for L.polyactis in addition to L.crocea,especially for their hybrid offspring lately developed and cultured in hatchery settings.Additionally,the methodology could inspire further research endeavors in microsatellite development at the genomic level for other non-model marine fish species that are in need of effective genetic markers.
Key words:Larimichthys crocea;Larimichthys polyactis;genome sequence;microsatellite DNA;cross amplification
1 Introduction
The advance of next generation high-throughput DNA sequencing (HTS) has revolutionized fishery and biological research.By employing the whole genome sequence,it is possible to develop large panels of genetic markers with relative ease,especially microsatellite DNA loci(microsatellites)[1]and single nucleotide polymorphisms(SNPs)[2-3].Microsatellites also known as simple sequence repeat loci (SSRs)are widely present and abundant in the genome of organism.The number of microsatellite repeat motifs varies greatly among individuals and populations within a species,making them an effective tool for population genetic assessment[4].They are co-dominant complying with the Mendelian inheritance,allowing for identifying heterozygosity,and also selectively neutral,and thus are powerful for the studies of genetic mapping,parentage assignment and conservation biology[5-7].
The large yellow croaker,Larimichthys crocea is an important commercial drum species (Order:Perciformes;Family:Sciaenidae),native to the East and South China Seas.Since the 1970s,the productivity of its fishery has been in decline progressively due to overexploitation of resources and loss of spawning and wintering grounds off the Coast of China;in the 1990s,wild stocks of L.crocea were almost depleted[8,9].The small yellow croaker (L.polyactis),the sole sibling species of L.crocea is also an important commercial marine fish native to the Yellow-Bo Sea (i.e.Yellow Sea and Bo Sea) and East China Sea.It has experienced a severe decline since the 1970s due to anthropogenic factors such as overfishing and environmental degradation,in addition to ocean current and seawater temperature changes[10-11].This study aimed to identify a new large dataset of microsatellites based on the genome DNA sequence of L.crocea[12]and further tested selected tetra-nucleotide loci across samples of L.crocea and L.polyactis.
2 Materials and methods
We screened microsatellite repeat motifs in the genome of L.crocea[12]using the MIcroSAtellite identification tool(MISA) v1.9.1 program[13].The minimum number of repeats was 10 for mono-nucleotide,six for di-nucleotide,and five for tri-,tetra-,penta-,and hexa-nucleotide microsatellites.We designed primer sets for tetranucleotide microsatellites following default criteria in PRIMER 3[14],and selected a set of the loci for further characterization in samples of L.crocea and L.polyactis.We labeled the forward primer of each microsatellite at the 5′ end with a fluorescent phosphoramidite,i.e.6FAM,HEX,ROX,or TAMARA (Applied Biosystems,USA).
We tested polymorphisms for a subset of the tetra-nucleotide microsatellites on 21 large yellow croaker and three small yellow croaker samples.YQ1-5 were fish larvae collected using trawl nets from Yueqing Bay,Yueqing,Zhejiang Province.At the point of capture,they were immediately immersed in 95% ethanol(laboratory grade).The larval samples were identified using mitochondrial Cytochrome c Oxidase subunit I (CO I) DNA barcoding[15].FD1-9 were same-generation juvenile fish spawned by a cultured stock of three pairs of males and females from a hatchery facility in Shacheng,Fuding,Fujian Province.Fin clips were taken from tails of eugenol anaesthetized fish and preserved in 95% ethanol (laboratory grade);ZJ1-10 were adult fish obtained from a local market in Naozhou Island,Zhanjiang,Guangdong Province.No fish were alive at the point of collection followed by preservation with 95% ethanol(laboratory grade).The juvenile and adult samples were identified using morphology[16].Detailed information for the samples was displayed in Table 1.

Tab.1 Information of L.crocea and L.polyactis samples used in this study,i.e.sample code,species identity,collection location including longitude and latitude,and sample size(n)
Genomic DNA of all samples was extracted using the OMEGA kit (Omega Bio-tek Inc USA).The concentration of DNA was quantified using 1.5% agarose gel electrophoresis.DNA samples were stored in a -20 °C freezer for later use.
We amplified the microsatellite loci via polymerase chain reaction (PCR).Each 20 μL PCR contained:2×Taq Mix (Promega USA),0.25 μ mol·L-1labeled forward primer,0.50 μ mol·L-1unlabeled reverse primer,and 10-100 ng DNA template.PCR amplification used following conditions:96 ℃for 4 minutes,followed by 35 cycles of 95 ℃for 30 seconds,52-55 ℃for 40 seconds,and 72 ℃for one min,ending with an extension of 72℃for 7 minutes.PCR products showing positive amplification were sent for fragment length analysis with ABI 3 100 (Applied Biosystems,USA) genetic analyzer at Genewiz (Suzhou,PR China).
We computed the allelic frequency in CONVERT 1.2[17],the polymorphic information content(PIC)in PIC_CALC online[18],and the number of alleles (NA),number of effective alleles (NE),observed heterozygosity (HO) and expected heterozygosity (HE) in POPGEN 1.32[19].
3 Results
We identified a total of 499 638 microsatellite loci across the 47 Mb L.crocea genome (data available in Table S1).The microsatellite type of di-nucleotide is most abundant,accounting for 49.66% of the total number of microsatellites,followed by the mono-(36.89%),tri (8.88%) and tetra-nucleotide (3.67%).Table 2 displayed statistical information of number and proportion of each specific type of microsatellite.
We developed primers sets for a total of 10 261 tetra-nucleotide microsatellite loci(data available in Table S2) and selected 32 of the loci for further characterization.We evaluated polymorphisms for 22 of the microsatellites that each showed amplification of no more than two alleles and/or less than 10% missing data across 21 L.crocea and 3 L.polyactis samples.Information for primer sets and Tmvalues,and for product sizes scored in base pairs and statistics of genetic parameters were shown in Table 3 and Table 4,respectively.In L.crocea,a total of 220 alleles were scored with an average of NA,10±5.58 alleles per locus.Lcr-09 and Lcr-10 had the greatest number of alleles,18 with the range of allele size scored in 227-341 bp and 178-342 bp,respectively;Lcr-08 had the least number of alleles,2 with the range of allele size scored in 251-255 bp;NE,HO,HEand PIC averaged over loci are 5.40±3.68,0.64±0.25,0.71±0.22 and 0.67±0.22,respectively.
Cross-species amplification in L.polyactis yielded 66 alleles in total for the 22 microsatellites with an average of NA,3±1.51 alleles per locus.Lcr-12 had the largest number of alleles,6 with the range of allele size scored in 191-319 bp;Lcr-07,Lcr-08,Lcr-09,and Lcr-18 each had only one allele,i.e.251,241,223 and 231,respectively;NE,HO,HEand PIC averaged over loci are 2.65±1.40,0.52±0.38,0.60±0.34 and 0.45±0.26,respectively.
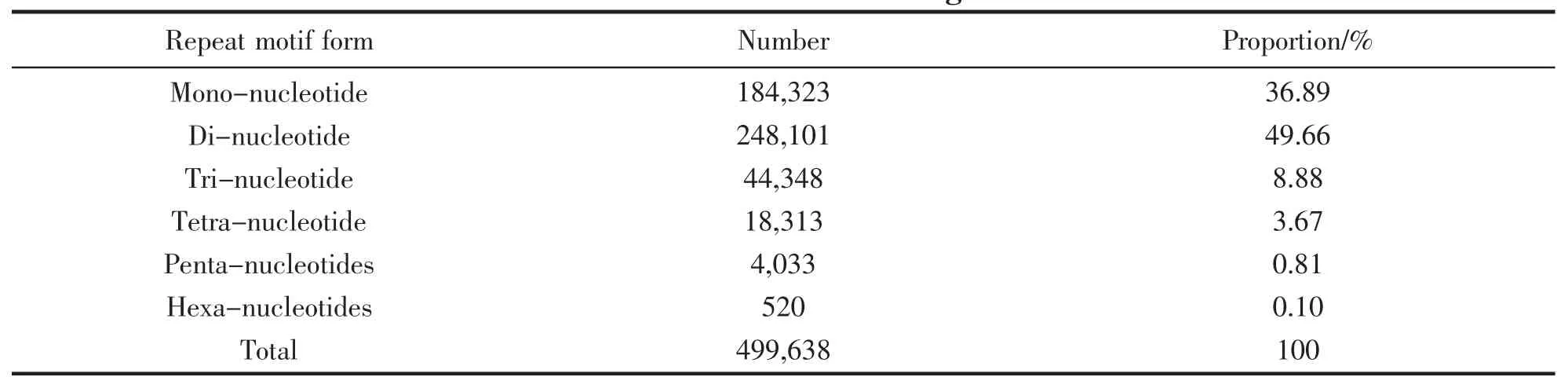
Tab.2 Statistical information of number and proportion for each specific type of microsatellite identified from the L.crocea genome

Tab.3 Information for primer set,repeat motif and Tm value of 22 tetra-nucleotide microsatellite loci tested for L.crocea and L.polyactis

Tab.4 Product sizes (allele range) scored in base pairs for 22 tetra-nucleotide microsatellite loci tested in L.crocea and L.polyactis and statistics of genetic parameters that include the number of alleles (NA),number of effective alleles (NE),observed heterozygosity (HO),expected heterozygosity (HE)and polymorphic information content (PIC).NA:not applicable.
4 Discussion
Understanding genetic variability in species that are rare or threatened is of particular and broad interest[20-21].Despite that microsatellite markers were previously developed in L.crocea,e.g.,from genome[22]and transcriptome resources[23],this study identified a new large dataset of microsatellites for L.crocea and L.polyactis based on the latest updated genome sequence data of L.crocea[12].In this pioneer study,the sample sizes of both species are fairly small,in particular for L.polyactis;therefore,we are unable to draw final conclusions until analyses with larger sample sizes are conducted.Nonetheless,these microsatellite DNA markers could crossamply and thus benefit advanced studies in population genetics and stock assessment for L.polyactis in addition to L.crocea,especially for their hybrid offspring lately developed and cultured in hatchery settings[24].The data could also facilitate future germplasm conservation and marker-assisted selection breeding programs for these two important commercial croaker species.In addition,the methodology could inspire further research endeavors in microsatellite development at the genomic level for other non-model marine fish species that are in need of effective genetic markers.
Acknowledgements
The study was supported by the Zhejiang Provincial Natural Science Foundation(LY16D060002),State Key Laboratory of Genetic Resources and Evolution of Kunming Institute of Zoology,Chinese Academy of Sciences(GREKF16-03),Zhejiang Science and Technology Innovation Program for College Students-Xinmiao Talent Program (18857090309),and Open Foundation from Marine Sciences in the First-Class Subjects of Zhejiang Province.We thank Michael Reuscher (Texas A&M University -Corpus Christi) and Autaipohn Kaikaew (Burapha University) for their critical and insightful comments on the manuscript.
Compliance with ethical standards
Conflict of interest:The authors declare that they have no conflict of interest.
Supplementary data are available
Table S1 Supporting information about the 499,638 SSRs (microsatellite markers) identified from the genome sequence of L.crocea,including chromosome,scaffold identity,SSR type,SSR repeat motif and specific position (start and end) in reference to the L.crocea genome.
Table S2 Supporting information for the primer sets of 10,261 tetra-nucleotide SSRs (microsatellites) identified from the genome sequence of L.crocea (five pairs for each SSR),including chromosome,SSR type,SSR repeat motif and specific position(start and end) in reference to the L.crocea genome.Note:22 microsatellite loci tested in this study.

