Knockout of ebony gene leads to melanin pigmentationin the rice stem borer, Chilo suppressalis(Lepidoptera: Crambidae)
SUN Hao, HUANG Jing-Mei, LIU Yan, GE Wen-Chao, WANG Shuai,YANG Feng-Xia, GAO Cong-Fen, WU Shun-Fan,*
(1. College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China;2. State & Local Joint Engineering Research Center of Green Pesticide Invention and Application, Nanjing 210095, China)
Abstract: 【Aim】 The rice stem borer, Chilo suppressalis, is a destructive rice pest in China and other Asian countries. However, due to lack of genetic tools, the functional genomic studies in C. suppressalis are seriously constrained. The aim of the study is to use a marker gene, ebony, to establish a gene editing system based on CRISPR/Cas9 technology in C. suppressalis. 【Methods】 With the amino acid sequences of Bombyx mori ebony protein as a query, the putative C. suppressalis ebony gene was obtained on its genomic database by the TBLASTN program. The full-length cDNA of ebony gene of C. suppressalis was cloned by PCR and subjected to bioinformatical analysis. The expression patterns of Csebony at different developmental stages (egg, larval, pre-pupal, pupal, and male and female adult stages) and in multiple tissues (head, epidermis, fat body, gut, and Malpighian tubules) of the 4th instar larvae of C. suppressalis were analyzed by qRT-PCR. Finally, we performed targeted knockout of ebony gene in C. suppressalis by microinjecting the ribonucleoprotein complexes specific guide RNA/Cas9 protein into the newly laid eggs within 2 h based on the CRISPR/Cas9 gene editing technology. 【Results】 The full-length cDNA of Csebony gene (GenBank accession no.: MZ846208) of C. suppressalis was cloned. It contains a 2 586 bp ORF encoding 861 amino acids, with the molecular mass of 9.5 kD and theoretical isoelectric point of 5.10. Csebony has no signal peptide sequence at the N-terminus. Domain analysis showed that Csebony has three conserved domains. Phylogenetic analysis indicated that Csebony is most closely related to Ostrinia furnacalis ebony. The qRT-PCR results showed that Csebony was highly expressed in the pupal stage and head. Knockout of Csebony caused melanin pigmentation in larvae, pupae, and adults of C. suppressalis. 【Conclusion】 The results showed that Csebony is involved in regulating cuticle pigmentation of C. suppressalis, and CRISPR/Cas9-based genome editing technology is effective in C. suppressalis. We can use visible marker gene to establish CRISPR/Cas9-based genome editing system in non-model organisms, so as to offer a valuable genetic tool for the study of functional genomics in C. suppressalis.
Key words: Chilo suppressalis; gene editing; CRISPR/Cas9; ebony; marker gene; phenotype; off-target effect
1 INTRODUCTION
The striped rice stem borer,Chilosuppressalis, is one of the most destructive economically important pests on rice (Savaryetal., 2019). Chemical control is an important way to restrict the occurrence and damage ofC.suppressalis(Gaoetal., 2013; Wuetal., 2014). However, the long-term, large-scale, and rampant use of chemical insecticides has resulted in development of resistance inC.suppressalisto commonly used insecticides (Yaoetal., 2017; Huangetal., 2020). Thus, new pest management strategies have to be developed for control of rice stem borer.
The Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 (CRISPR/Cas9), as the next-generation gene editing technology after zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) (Urnovetal., 2010; Milleretal., 2011), has been regarded as an efficient and powerful tool to explore gene functions in human cells (Congetal., 2013; Malietal., 2013), zebrafish (Hwangetal., 2013), mice (Wang Hetal., 2013), andDrosophilamelanogaster(Bassettetal., 2013). In addition, CRISPR/Cas9 can be used as a functional genomic tool in deciphering insecticide resistance and has shown great potential for the development of novel pest management strategies (Homem and Davies, 2018; Dourisetal., 2020). For example, Wangetal.(2020) introduced the I4790M mutation in the ryanodine receptor (RyR) ofPlutellaxylostellaand validated the role of this mutation in conferring moderate levels of resistance to flubendiamide. In mathematical simulations and cage experiments, CRISPR/Cas9-mediated gene drive systems can suppress the population of the malaria vectorAnophelesgambiaeby targeting genes related to female fertility (Hammondetal., 2016; Kyrouetal., 2018).
A visible marker gene that allows rapid and reliable identification of knockout cases is very important for the further development of gene editing methods in non-model insects (Bietal., 2019; Zhuetal., 2020). Theebonyencodes the enzyme N-β-alanyl dopamine synthetase, which catalyzes the synthesis of N-β-alanyl dopamine (NBAD) from β-alanine and dopamine (Fukushi and Seki, 1965; Wright, 1987; Hovemannetal., 1998). The biosynthetic pathways are essential for the melanin synthesis (Boryczetal., 2002; Wittkoppetal., 2002). The function ofebonygene has been demonstrated in other insect species to be involved in pigmentation at different developmental stages (Wittkoppetal., 2002; Futahashietal., 2008; Lietal., 2015; Lietal., 2017; Ryleeetal., 2018; Bietal., 2019).
In this study, we analyzed both the molecular features and expression patterns ofebonygene ofC.suppressalis(Csebony). To develop genetic tools inC.suppressalis, we delivered the ribonucleoprotein complexes into the fresh eggs ofC.suppressalisresulting in the disruption ofCsebonyinvivo. Experimental data showed thatCsebonygene plays a significant role in regulating the body coloration of larvae, pupae, and adults. Using a marker gene,ebony, this study established an efficient CRISPR/Cas9-based gene editing method inC.suppressalisfor functional genomic studies.
2 MATERIALS AND METHODS
2.1 Insects
The wild type strain ofC.suppressalis(named BJ18) was provided by Dr. HAN Lan-Zhi of Chinese Academy of Agricultural Sciences in 2018 and reared in the laboratory. Larvae were fed with rice seedlings and placed in a constant temperature incubator with a temperature of 28±1℃ and a photoperiod of 16L∶8D. Pupae were collected in a sterilized plastic petri dish for pupation. After emergence, moths were transferred to insect cages with a relative humidity of 85%-90% and fed with 10% honey water as nutrition.
2.2 Cloning of ebony from C. suppressalis
With the amino acid sequences ofBombyxmoriebony protein as a query, the putativeebonygene ofC.suppressaliswas identified by searching its genomic database (Maetal., 2020) using TBLASTN program. Total RNA was extracted from the 4th instar larvae using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). The cDNA was synthesized by HiFiScript cDNA Synthesis Kit (Proteinssci, Shanghai) following the manufacturer’s protocol. The cDNA fragment ofCsebonywas amplified by PCR, the primers were designed using Primer Premier 5 software and listed in Table 1. The specificity of the primers was examined using NCBI Primer-blast tool (http:∥www.ncbi.nlm.nih.gov/tools/primer-blast/). Each 25 μL PCR reaction included 1 μL of forward primer (10 μmol/L), 1 μL of reverse primer (10 μmol/L), 2 μL of cDNA, 12.5 μL 2×Phanta Max Master Mix (Vazyme, Nanjing) and 8.5 μL ddH2O. Using high-fidelity enzyme to carry out PCR reaction, the specific reaction procedure was as follows: 95℃3 min; 35 cycles of 95℃15 s, 54℃ 15 s, 72℃ 3 min; 72℃10 min. The PCR amplification products were cloned into the pClone007 Blunt Simple vector (TSINGKE, Beijing) and then sequenced using the 3 730 XL DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA).
2.3 Bioinformatical analysis of Csebony
The molecular weight, protein length and isoelectric point (pI) ofCsebonywere predicted by an online software Expasy (http:∥web.expasy.org/protparam/). The SignalP 5.0 Server (http:∥www.cbs.dtu.dk/services/SignalP) was used for prediction of the signal peptide. The phylogenetic tree was constructed by MEGA-X using maximum likelihood method (Kumaretal., 2018). Bootstrap values were calculated after 1 000 replicates to test the credibility of the evolutionary tree branch.
2.4 Developmental and tissue expression profiling of Csebony gene
qRT-PCR was performed to determine the expression profiles ofCsebonyat different developmental stages (1, 4 and 6 d-old eggs, day-1-4 1st instar larvae, day-1-3 2nd-4th instar larvae, day-1-2 5th instar larvae, day-1 6th instar larva, pre-pupa, day-1, 4 and 7 pupae, and male and female adults) and in multiple tissues of the 4th instar larvae (head, epidermis, fat body, gut, and Malpighian tubules) (30 larvae were sampled for each biological replicate). Each biological replicate includes 100 eggs and six individuals of larvae, pre-pupae, pupae, and adults. Primers were designed using Primer 3 Input (https:∥primer3.ut.ee/) based on the sequence cloned in section 2.2 to amplify a 191 bp fragment and are listed in Table 1. Total RNA was isolated from above samples using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized using HiScript®II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme, Nanjing) according to the manufacturer’s protocol. PCR reactions (20 μL) containing 4 μL of cDNA (80 ng/μL), 10 μL of AceQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing), 0.2 μmol/L of each primer and 5.2 μL ddH2O. Samples were run on Applied Biosystems 7 500 Fast following the manufacturer’s protocol. Three independent biological replicates with four technical replicates were performed for each sample. Data were calculated according to the 2-ΔΔCtmethod using the geometric mean of two selected reference genesactinA1 andG3PDHfor normalization (Table 1) (Vandesompeleetal., 2002).
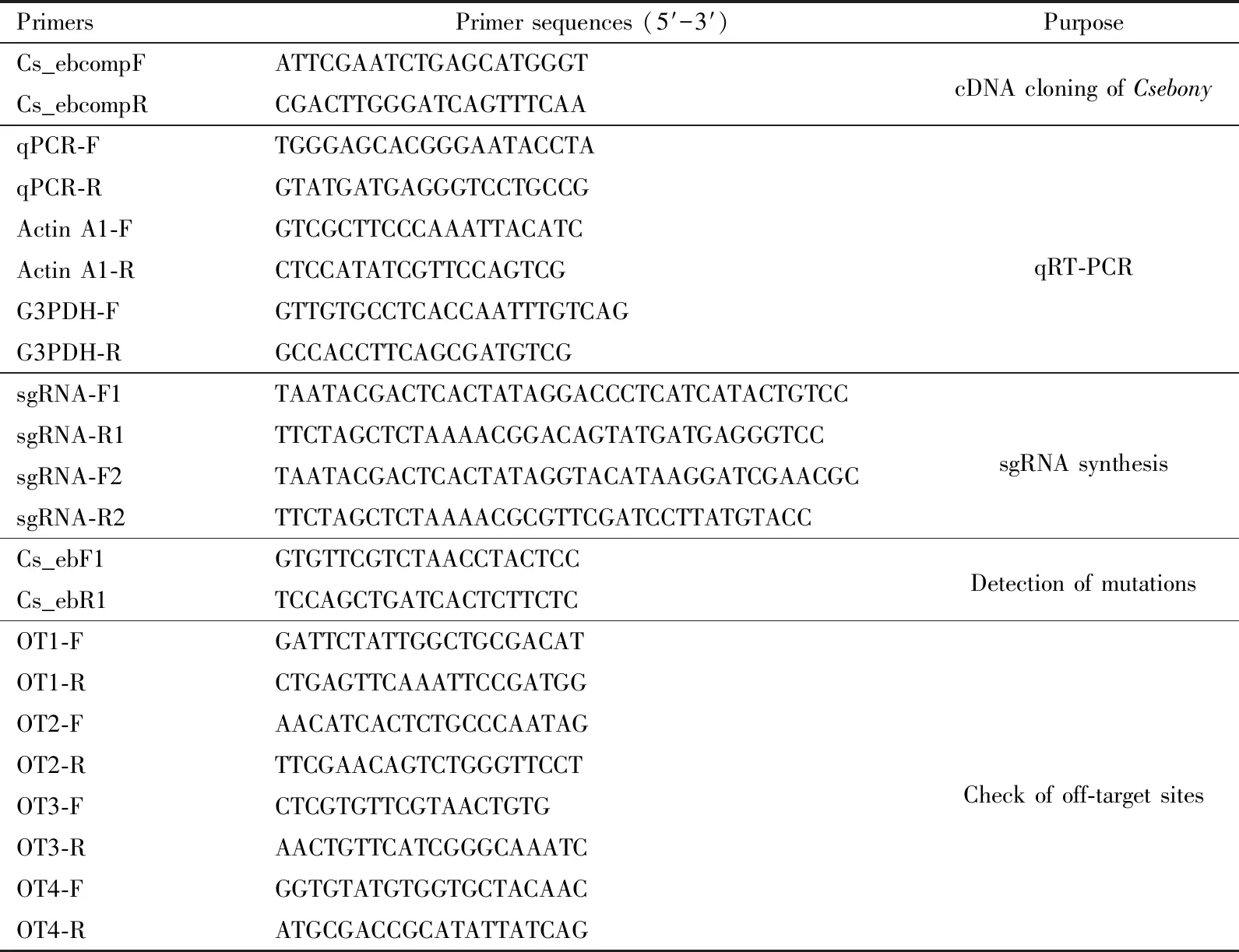
Table 1 Primers used in this study
2.5 Microinjection of sgRNA and Cas9 protein
The selection of sgRNA target sites was based on the criterion: 5′-GG-(N) 18-NGG-3′ (Wang Yetal., 2013). Thus, a 20 bp targeting site (5′-GGACCCTCATCATACTGTCC-3′) located on exon 5 and another targeting site (5′-GCGTTCGATCCTT ATGTACC-3′) located on exon 6 were selected to design the sgRNA via the CRISPR gRNA Design tool-ZiFiT 4.2 (http:∥zifit.partners.org/ZiFiT/Disclaimer.aspx). Two sgRNA templates were generated using four specific oligonucleotides shown in Table 1. The sgRNAs were synthesizedinvitrousing the GeneArtTMPrecision gRNA Synthesis Kit (Thermo Fisher Scientific, Vilnius) according to the instructions of manufacturer. Purified sgRNAs were stored at -80℃ until use. The Cas9 protein (GeneArtTMPlatinumTMCas9 Nuclease) was purchased from Thermo Fisher Scientific (Shanghai). The fresh eggs ofC.suppressaliswere cut from the rice leaves within 2 h after laying, glued on the glass slide with double-sided adhesive tape, and injected using the microinjection system (InjectMan NI 2 microinjection system, Eppendorf, Hamburg) with a mixture containing 300 ng/μL Cas9 protein (GeneArtTMPlatinumTMCas9 Nuclease) and 150 ng/μL of each sgRNA. We injected the same volume of water as the negative control. Injected eggs were incubated at 28±1℃ and 60%±5% RH.
2.6 Mutagenesis analysis
To determine whether CRISPR/Cas9 successfully knocked outCsebonyin section 2.5, the genomic DNA fragments ofCsebonyflanking the target regions were amplified (Table 1). Genomic DNA was isolated from individual newly hatched larva using AxyPrep Multisource Genomic DNA Miniprep Kit (Axygen, Hangzhou) following the manufacturer’s instructions. The PCR amplification products were run on a 1.5% agarose gel to verify that the mutation occurred, and then subcloned into the pMD-19T vector (TaKaRa, Dalian) and sequenced to determine the exact mutation types. Ten clones were randomly chosen for Sanger sequencing to examine insertions and deletions sequenced.
2.7 Off-target effect analysis
To test for off-target effects, we applied the CasOT (Xiaoetal., 2014) to predict potential off-target sequences in the rice stem borer genome (Maetal., 2020). According to previous study (Renetal., 2014), off-target effects did not occur in regions with three or more nucleotide mismatches to sgRNAs. We did not find any sequences with less than three nucleotide mismatches with sgRNA. Nevertheless, we selected the top two off-target sites corresponding toCsebonysgRNA1 and sgRNA2, respectively (Table 2). We randomly chose three individuals from G0 mutant larvae to examine two off-target sites with PCR analysis. The DNA fragments spanning potential off-target sequences were amplified by specific primers (Table 1) and sequenced.
2.8 Data analysis
All data were presented as mean ±SD. One-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test was used for multiple group comparisons.P<0.05 was considered significant statistically. All statistical analyses were performed using GraphPad Prism 8 Software (GraphPad, San Diego, CA, USA).
3 RESULTS
3.1 Identification and phylogenetic analysis of Csebony
Direct sequencing of the PCR product showed that the cDNA ofCsebonygene (GenBank accession number: MZ846208) contains a 2 586 bp ORF encoding 861 amino acids with a molecular weight of 9.5 kD and an isoelectric point of 5.10. Csebony protein is encoded by 16 exons separated by introns of various lengths without signal peptide sequence at the N-terminus. The amino acid sequence alignments of Csebony with ebony proteins of other insects revealed that these proteins contain three major domains: The acyl-activating enzyme (AAE) consensus motif near the N-terminus, [LIVMFY]-X-X-[STG]-[STAG]-G-[ST]-[STEI]-[SG]-X-[PASLIVM]-[KR] (where X is any amino acid), adenosine monophosphate (AMP) binding sites scattered in the sequence, and phosphopantetheine attachment site (PP binding) (Fig. 1). The phylogenetic tree showed that Csebony is most closely related toOstriniafurnacalisebony. A closely related clade also contains ebony protein of other lepidopteran insects, suggesting a conserved function (Fig. 2).
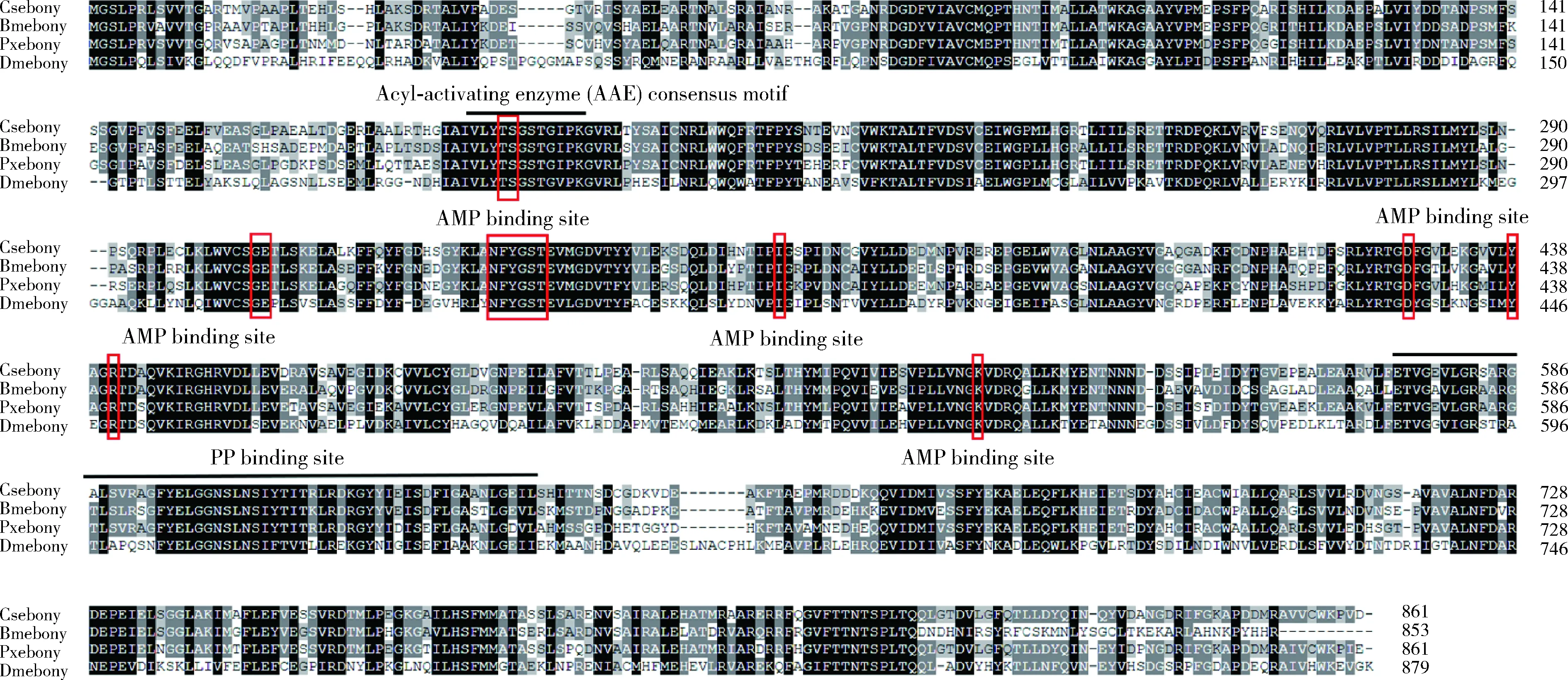
Fig. 1 Amino acid sequence alignment of ebonyOrigin species of ebony proteins and their GenBank accession numbers: Csebony: Chilo suppressalis, AKL78853.1; Pxebony: Papilio xuthus, BAE43845.2; Bmebony: Bombyx mori, BAH11147.1; Dmebony: Drosophila melanogaster, ABO27276.1.
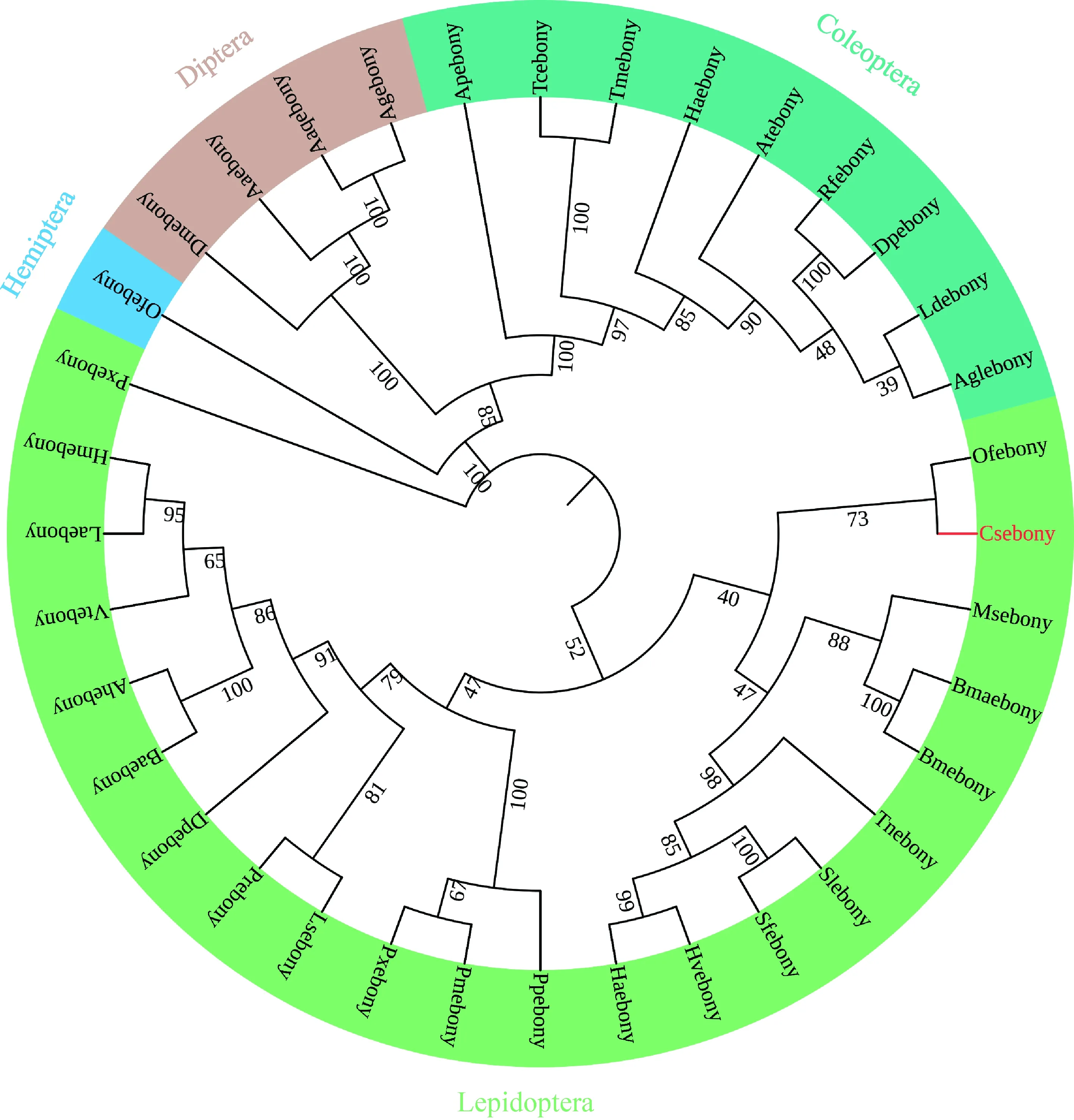
Fig. 2 Phylogenetic analysis of Csebony of Chilo suppressalis with the ebony proteins from other insect speciesconstructed by maximalum likelihood method based on amino acid sequence (1 000 replicates)Origin species of ebony proteins and their GenBank accession numbers: Csebony: Chilo suppressalis, AKL78853.1; Bmebony: Bombyx mori, BAH11147.1; Haebony: Helicoverpa armigera, XP_021196746.1; Pxebony: Plutella xylostella, XP_011549000.1; Sfebony: Spodoptera frugiperda, XP_035441626.1; Msebony: Manduca sexta, XP_030033133.1; Bmebony: Bombyx mandarina, XP_028043469.1; Slebony: Spodoptera litura, XP_035441645.1; Ofebony: Ostrinia furnacalis, XP_028157131.1; Pxebony: Papilio xuthus, BAE43845.2; Pmebony: Papilio machaon, BAJ07590.1; Ppebony: Papilio polytes, BAJ07596.1; Hmebony: Heliconius melpomene malleti, ADU32896.1; Dpebony: Danaus plexippus plexippus, OWR52883.1; Ahebony: Aphantopus hyperantus, XP_034837374.1; Tnebony: Trichoplusia ni, XP_026745827.1; Vtebony: Vanessa tameamea, XP_026492433.1; Hvebony: Heliothis virescens, PCG65717.1; Laebony: Limenitis arthemis astyanax, QHN70668.1; Baebony: Bicyclus anynana, XP_023943105.1; Prebony: Pieris rapae, XP_022123243.1; Lsebony: Leptidea sinapis, VVD05713.1; Tmebony: Tenebrio molitor, QGQ59667.1; Tcebony: Tribolium castaneum, XP_008197905.1; Atebony: Aethina tumida, XP_019873373.1; Agebony: Anoplophora glabripennis, XP_018568645.1; Ldebony: Leptinotarsa decemlineata, ATB56363.1; Dpebony: Dendroctonus ponderosae, ERL87248.1; Haebony: Harmonia axyridis, QNH91384.1; Rfebony: Rhynchophorus ferrugineus, KAF7285868.1; Apebony: Agrilus planipennis, XP_025833498.1; Ofebony: Oncopeltus fasciatus, AMW91811.1; Dmebony: Drosophila melanogaster, ABO27276.1; Aaebony: Aedes aegypti, XP_001651463.2; Agebony: Anopheles gambiae, EAA03788.5; Aaqebony: Anopheles aquasalis, JAB00393.1.
3.2 Developmental and tissue expression profiles of Csebony
Csebonywas widely expressed at all developmental stages, with the high expression level at the prepupal and pupal stage and the lowest expression level in the 1 d-old egg stage (Fig. 3: A). In addition, the mRNA level ofCsebonywas higher before and after molting, and was lower in the day-2 larvae of various instars (Fig. 3: A). Different tissues, including fat body, head, epidermis, gut, and Malpighian tubules, were dissected from the day-1 4th instar larvae ofC.suppressalis.Csebonywas mostly expressed in the head compared with other examined tissues (Fig. 3: B).
3.3 Phenotypes induced by knockout of Csebony
Seven days after microinjection, 46.1% (507 out of 1 100) of eggs were successfully hatched to the 1st instar larvae. The study showed that, in theCsebony-sgRNA treatment, the coloration of larval abdominal epidermis of the mutant was deeper than that of the wild type (Fig. 4: B). This phenotype was observed in the 2nd instar larvae. Approximately, 73.9% (374/509) of hatched larvae were detected with a mutant phenotype ofCsebonygene. In addition, the color of head capsule and pronotum ofebonymutants was darker than that of the controls (Fig. 4: A, C).
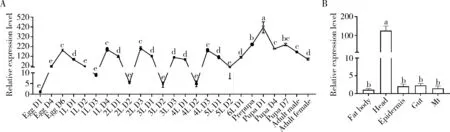
Fig. 3 Relative expression levels of Csebony at different developmental stages (A) and in various tissuesof the 4th instar larvae (B) of Chilo suppressalis determined by qRT-PCREgg D1, D4, D6: 1, 4 and 6 d-old eggs, respectively; 1L D1-4: Day-1-4 1st instar larvae, respectively; 2L D1-3: Day-1-3 2nd instar larvae, respectively; 3L D1-3: Day-1-3 3rd instar larvae, respectively; 4L D1-3: Day-1-3 4th instar larvae, respectively; 5L D1-2: Day-1-2 5th instar larvae, respectively; 6L D1: Day-1 6th instar larva; Pupa D1, D4, D7: Day-1, 4 and 7 pupae, respectively. Mt: Malpighian tubules. The gene expression levels at different developmental stages and tissues were normalized to those in the Egg D4 and fat body, respectively. Data in the figure are mean±SD. Different lowercase letters above bars indicate significant differences in the gene expression level among different developmental stages and tissues (P<0.05) as determined by one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test.

Fig. 4 Phenotypic changes of Chilo suppressalis caused by knocking out Csebony gene by CRISPR/Cas9 systemA: Knockout of Csebony gene leads to color deepening of pronotum and head capsule. Scale bar=1 mm. B: Knockout of Csebony gene induces pigmentation of abdominal epidermis. Scale bar=1 mm. C: Magnified image of head capsule (A). Scale bars=0.5 mm. D: Melanized puparium in Csebony mutants. Scale bar=2 mm. E: Mutant adults exhibited deep pigmentation. Scale bars=1 mm.
We observed that knockout ofCsebonyleads to melanization of the puparium. Two abnormal phenotypes, spot mutants and integral mutants, were also presented in theCsebonymutants (Fig. 4: D). It was believed that spot mutants were chimera and more mutant cells exited in the integral mutants. After emergence, the coloration of the forewings, antennae, and leg of the mutant moths were also deeper than that of the wild type (Fig. 4: E).
3.4 Off-target (OT) detection in G0 generation mutant larvae
Totally four potential off-target sites were analyzed in three individuals randomly selected fromCsebonymutant larvae. The results showed a single peak in all the tested sequences (Fig. 5). Single peak suggested that body-color change was derived fromCsebonygene mutation rather than off-target effects.
4 DISCUSSION
Ebonyhas been extensively used as a marker gene for developing CRISPR/Cas9 method in the corresponding species (Lietal., 2017; Ryleeetal., 2018; Bietal., 2019). In this study, we successfully applied CRISPR/Cas9-mediated gene editing technology to study the function ofebonyin the non-model insectC.suppressalis. Firstly, we cloned the full-length cDNA ofebonygene inC.suppressalis.Secondly, we found thatCsebonywas expressed in all developmental stages and examined tissues with highly expressed in the pupal stage and head (Fig. 3). Thirdly, we synthesized two sgRNAs The off-target sites are noted as non-seed_seed-PAM, in which lowercase letters in the sequence represent mismatched nucleotides. Mismatch type: For example, A13 represents 1 mismatch in the seed sequence region and 3 mismatches in the non-seed sequence region. Mismatch all: The total number of mismatches.
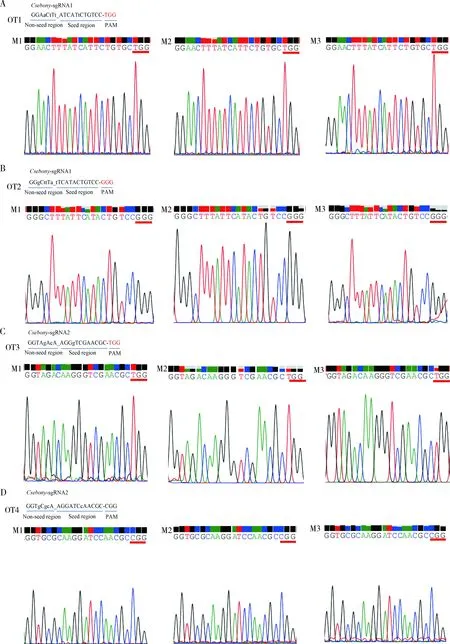
Fig. 5 Potential off-target sites of sgRNA of Csebony gene of Chilo suppressalis analyzed by DNA sequencingFor off-target site information, see Table 2.
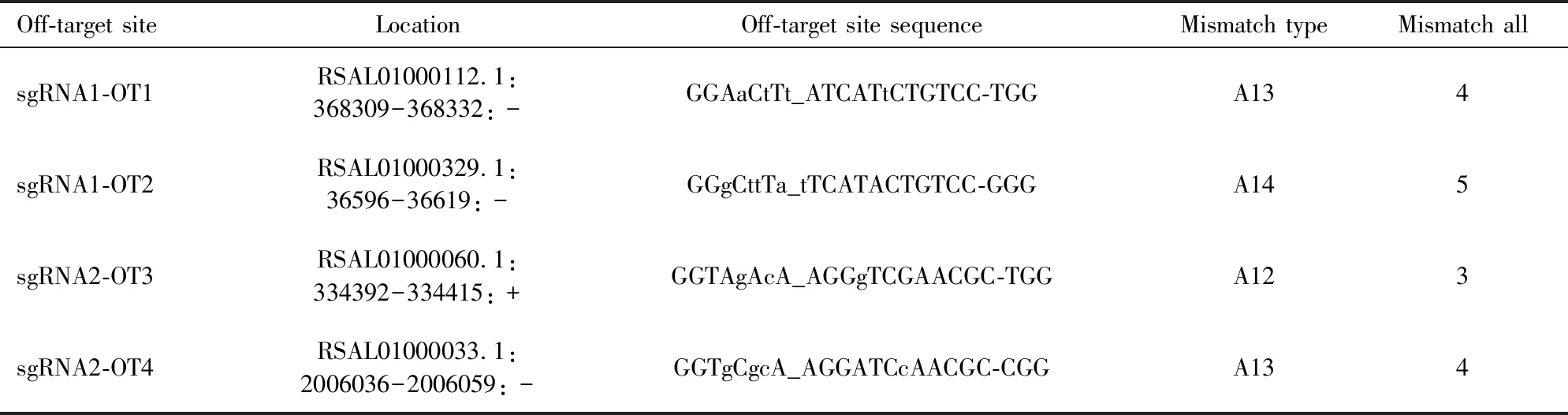
Table 2 Potential off-target (OT) sites of Csebony gene of Chilo suppressalis
which target on exons 5 and 6, respectively, and we injected a mixture of Cas9 and gRNA into fresh eggs that resulted in highly efficient editing (73.9%) of theCsebonylocus. In agreement with CRISPR/Cas9-mediatedebonymutants inS.litura(Bietal., 2019), the loss-function ofCsebonygene also affected the coloration of the puparium. In the adult stage, the forewings, leg, and antenna ofCsebonymutants showed deeper color than wild type (Fig. 4).
Csebonywas expressed at all developmental stages and highly expressed in the pupal stage (Fig. 3: A). The temporal expression pattern ofCsebonywas similar to the previous studies (Lietal., 2015; Bietal., 2019). InP.xuthus,P.machaonandS.litura,ebonywas expressed not only at larval and pupal stages but also at adult stage (Lietal., 2015; Bietal., 2019). In addition,Slebonywas expressed at the pupal stage, especially at the pre-pupal stage ofS.litura(Bietal.,2019). The tissue-specific expression pattern ofCsebonywas consistent with that previously described inS.litura(Bietal., 2019). TheebonymRNA was detected at relatively high levels in the head (Fig. 3: B), indicating thatebonymay play similar roles in tissue-specific larval color patterns. InP.xuthus, cuticle pigmentation occurred 14-16 h after head capsule slippage (HCS) andebonywas highly expressed at a few hours before molting. After the cuticle pigmentation stage, the expression ofebonywas declined (Futahashi and Fujiwara, 2005). Similarly, we found thatCsebonywas highly expressed in the last day larvae of various instars (a few hours before molting). After molting, the expression level ofCsebonywas gradually decreased.
Ebonywas involved in melanin biosynthesis by combining β-alanine with dopamine to form NBAD (Fukushi and Seki, 1965; Wright, 1987).Csebonymutant displayed enhanced cuticle pigmentation at different developmental stages. This phenotype is similar toebonymutant inP.xuthus(Lietal., 2015),Bombyxmori(Futahashietal., 2008), andDrosophila(Wittkoppetal., 2002). InP.xuthus, the previous study showed that coloration pattern of the epidermis of larvae changed dramatically after the 4th molting andebonywas strongly expressed in the reddish-brown area of the 5th instar larva (Futahashi and Fujiwara, 2005). Mutated 5th instar larvae exhibited enhanced melanic pigmentation in reddish-brown area, and adult mutants showed brown pigmentation across the body (Lietal., 2015). A similar result was reported inB.mori, loss-function ofebonyalleles caused a smoky phenotype of larvae and black pupa, suggesting thatebonyis involved in the pigment patterning (Futahashietal., 2008). In addition to lepidopteran insects, the same phenomena also appear in dipteran insectDrosophila. Theebonymutant was much darker than the wild type in the cuticle anterior to the stripe, the thorax, and wings (Wittkoppetal., 2002). Taken together, phenotypic analysis demonstrated thatebonyhas conserved functions in pigment biosynthesis among multiple species.
Mutagenesis efficiency is a direct reflection point of the CRISPR/Cas9 system. High efficiency of gene editing is obtained to produce more G0 generation mutants, providing more experimental insects for subsequent verification experiments. The mutation efficiency is determined by multiple factors, including the concentration of gRNA and Cas9 protein (Renetal., 2013), the delivery method of Cas9 protein (Kistleretal., 2015), the amount of gRNA (Geetal., 2016; Kaneetal., 2017),etc. After comprehensively considering the above factors, we delivered two sgRNAs and the Cas9 protein into the embryos at optimal concentrations. Mutation rate of G0 larvae was 73.9% approximately, much higher than those reported in other studies (Bietal., 2019). These results indicated that CRISPR/Cas9 system works very well inC.suppressalis. With the CRISPR/Cas9-mediated precise gene editing, we can facilitate functional genomics research onC.suppressalis. In addition, genetic manipulation based on CRISPR/Cas9 system is a promising strategy that will be valuable for pest control.

