Bacteroides utilization for dietary polysaccharides and their beneficial effects on gut health
Jioo Cheng, Jielun Hu, Fng Geng, Shoping Nie,*
a State Key Laboratory of Food Science and Technology, China-Canada Joint Lab of Food Science and Technology (Nanchang),Nanchang University, Nanchang 330047, China
b Key Laboratory of Coarse Cereal Processing (Ministry of Agriculture and Rural Affairs), School of Food and Biological Engineering,Chengdu University, Chengdu 610106, China
Keywords:
Bacteroides
Polysaccharides
Degradation
Short-chain fatty acids
Gut health
A B S T R A C T
Polysaccharide was a class of macromolecular substance with various bioactive functions. Gut symbiotic microorganisms could utilize the polysaccharides from various sources, thus have important impact on human health. Bacteroides represented one of the dominant colonizers in the human gut. The utilization of polysaccharide by Bacteroides was important for supporting the function and stability of gut microbiota.After the degradation of polysaccharides by Bacteroides, gut microbes could ferment the monosaccharides and oligosaccharides degraded from polysaccharides into some metabolites, such as short-chain fatty acids (SCFAs), amino acids, etc. Among the metabolites, the SCFAs could have beneficial effects on gut health. This review summarized the niches of Bacteroides among gut microbiota, and also described the gene clusters and membrane proteins involved in the utilization processes of polysaccharide by gut Bacteroides.SCFAs could act as energy substrates for intestinal epithelial cells, inhibit histone deacetylases and activate G protein-coupled receptors. In addition, the future perspectives in investigating new degradation pathways for polysaccharide, and using polysaccharides or their metabolites as therapeutic approaches for diseases mediated by the gut dysbiosis were also provided.
1. Introduction
Gut microbiota represented trillions of microorganisms located in the gut, which could be up to 1 000 different bacterial species [1,2].Bacteria among the gut microbiota could perform many crucial functions in the maintenance of human health, which were also connected to hormone system, immune system and other metabolic pathways in the host. Disordered gut microbiota has been testified to increase the risk of metabolic disorders-related diseases, such as diabetes,cardiovascular disease, inflammatory bowel disease and so on [3].Approximately, 4 × 1013bacteria were densely colonized in the gut,which mainly included Firmicutes and Bacteroidetes, as well as Proteobacteria, Actinobacterium, Verrucomicrobia [4]. Firmicutes and Bacteroidetes were the dominant groups among the gut microbiota.Firmicutes was the largest and most diverse bacterial phylum, which could account for about 70% of bacterial phylogenetic diversity in the gut microbiota [5]. However, Bacteroidetes, represented by some core genera (Bacteroides,Alistipes,ParabacteroidesandPrevotella),could produce a larger number of polysaccharide-degrading enzymes than Firmicutes [6].
Dietary nutrients could influence the composition and metabolism of gut microbiota. The majority of dietary nutritional components included the fats, proteins, and monosaccharides, which were absorbed in the small intestine and unable to be used by the microbial community which inhabited in the large intestine [7]. Therefore, the direct source of energy for the gut microbes could come from the indigestible polysaccharides (such as resistant starch and cellulose)in diet. Among the main genera of Bacteroidetes,Bacteroideswere considered as the effective degraders for polysaccharides. The strong polysaccharide utilization ability ofBacteroidescould provide suitable nutrients for other bacteria, and contribute to the symbiotic relationship of gut communities [8].
Polysaccharide was defined as a long-chain polymer, which was consisted of more than 10 monosaccharide molecules connected by glycoside bonds. The kinds of dietary polysaccharides were diverse,including plant-source polysaccharides ranging from the simplest amylose to the most complex rhamnogalacturonan II, as well as the animal-source polysaccharides such as chondroitin sulfate, hyaluronan and heparin. Moreover, microorganism-source polysaccharides were primarily prepared from edible fungi, yeast and unicellular algae such asSaccharomyces cerevisiae,Hericium erinaceusandFucus vesiculosus. It was showed that lots ofBacteroidesspecies owned polysaccharide utilization loci (PULs) which were gene clusters encoding polysaccharide-degrading systems [9,10]. Meanwhile, gut microbiota could degrade and ferment polysaccharides into SCFAs,carbon dioxide, hydrogen and other metabolites closely related to the host health [11,12]. More detailed information should be provided for comprehensively understanding thatBacteroidespresented in the gut ecology utilize the polysaccharides and their beneficial effects for the gut health. Therefore, we discussed the physiological functions ofBacteroidesspecies, and the polysaccharide utilization system located inBacteroides. In addition, this paper also focused on the impacts of the important polysaccharide metabolite, such as SCFAs, on regulating the gut inflammation and maintaining the gut immune homeostasis.
2. Gut Bacteroides
2.1 Bacteroides in gut microbial ecology
Bacteroidaleswere the most abundant gram-negative bacteria which colonized in the human gut at densities of 108-109CFU/g of the feces[13]andincluded over 30 species which were more closely related to each other than other bacteria [14]. Up to now,10Bacteroidesspecies in the human body have been identified,which mainly includedB. vulgatus,B. caccae,B. uniformisandB. thetaiotaomicron[15,16].Bacteroideshad powerful nutrients utilization capabilities, which were partly depended on the ecological network for polysaccharide utilization [17]. There were also different patterns of competition behavior amongBacteroidespopulation. One competition behavior was the secretion of antimicrobial proteins.For example,B. fragilis638R secreted a eukaryotic-like ubiquitin protein which mediated intraspecies antagonism, andB. uniformisCL03T00C23 potently inhibited the growth ofB. uniformisATCC 8492 [18,19]. Another competition behavior was to deliver toxic effectors by type VI secretion systems, which was widespread in gram-negative bacteria in order to inhibit their competitors and attackers [20]. The balance between the cross-species cooperation and the interference competition ensured the stability ofBacteroidesin gut microbiota ecology.
Microbial energy in our gut was largely from the food particles, as well as the intestinal mucous layer and the exfoliated epithelial cells.These multi-directional means of obtaining the nutrients could help theBacteroidesto regulate its own genome when gut environment changed, which would also produce the adaptive enzymes. Among those, hybrid two-component signal transduction systems (HTCSs)and extracytoplasmic function sigma (ECF-σ) factors were the markedly expressed proteins involved in environmental sensing and transduction of the signal [21]. The phenomenon in which gut commensal bacteria respond to environmental cues was the presence of HTCSs. HTCSs combined the structure and function of core proteins from prototypic two-component systems into a single protein.One was a histidine protein kinase and the other was a response regulator protein. When an external signal acted on the ligand binding region outside the membrane, signal recognition promoted the binding and hydrolysis of ATP (adenosine triphosphate), and transferred the phosphate group of ATP to the histidine site on the kinase, which would cause it to the autophosphorylation. The phosphorylated sensor was served as phosphoryl donor to modify its cognate regulator,which was a DNA-binding transcriptional regulator [21]. Meanwhile,ECF-σ factors were defined as a class of environmentally responsive transcriptional regulators which owned divergent in sequence related to most other σ factors [22]. After receiving the stimulus from gut environment, ECF-σ factors were then released, and could bind to RNA polymerase to stimulate the transcription [23]. The multigene cluster related toB. thetaiotaomicronstarch utilization system (Sus) was consisted of the upstream gene encoding an ECF-σ factor. When environmental stimuli occurred, the expression of the ECF-σ factor was affected, which was accompanied by the expression or suppression of Sus [24]. Therefore, the nature of timely self-regulation ofBacteroidesafter sensing the stimulation factors in the environment could be recognized, which was also the fundamental reason for their existence as the dominant bacteria in the gut.
2.2 Physiological functions of Bacteroides
Bacteroidesspecies were largely studied for the regulatory effects on the host, which have been shown to accelerate the angiogenesis in the intestinal mucosa, improve the host immunity, and maintain the gut microecological balance (Table 1).
B. fragilisNCTC9343 alleviated DSS-induced colitis by increasing the anti-inflammatory cytokines IL-10 expression and suppressing the proinflammatory-related TNF-α and IL-6 expression[25]. The capsular polysaccharide in non-toxigenicB. fragilisNCTC9343 was able to reduce the bacteria-driven chronic colitis and inhibit the tumor development [26]. Moreover, the inhibitory role ofB. fragilisNCTC9343 treatment on the colon tumorigenesis had been proved by downregulating the expression of C-C chemokine receptor 5 in the gut [27].B. fragilisZY312 obtained from the healthy infant feces could protect the immune cells from the damage by the food-borne pathogenVibro parahaemolyticus[28].B. thetaiotaomicroncould serve as an environmental agent which was assigned the responsibility for regulating the development of intestinal angiogenesis via Paneth cells[29]. The cell surface-exposed lipoproteins (BtuG) ofB. thetaiotaomicronVPI-5482 could help the host to acquire the cyanocobalamin in the gut [30]. The antigen fromB. thetaiotaomicronVPI-5482 was able to restrain the activation of a CD4+T cell hybridoma via modulating colonic T cell responses [31].
One facet from the many dialogues betweenBacteroidesand the host was the induction of natural antibodies to participate in immunosurveillance. The dietary supplementation ofB. xylanisolvensboosted the level of natural Thomsen-Friedenreich alpha (TFα)-specific IgM serum antibodies [32]. In addition,B. vulgatusandB. doreipossessed intervention potentials of atherosclerotic lesion formation due to effectively improving endotoxemia,decreasing the production of gut microbial lipopolysaccharide and markedly inhibiting proinflammatory immune response [33].Some researchers used gene editing to introduce targeted gene fragments into the genome ofBacteroidesstrains so that better exhibition of modulatory effects can be achieved. By constructing plasmids containing inulin utilization genes and using inulin as a screening marker, genetic manipulation was realized for ErmR/TetRBacteroides/Parabacteroidesstrains that could not use inulin by themselves [34]. Some other researchers inserted sequence encodingTGF-β1gene into downstream of the xylanase promoter in the xylan operon ofB. ovatus. Therefore, the reconstructedB. ovatussecreted high bioactive dimeric TGF-β under the stimulation of xylan, and clinically relieved colitis [35].
Beneficial host effects ofBacteroidesdid not occur only in the gut. It was demonstrated thatB. acidifacienscould reduce the body weight and the fat mass by decreasing the intestinal dipeptidyl peptidase-4 (DPP-4), increasing the expression level of peroxisome proliferator-activated receptor alpha (PPARα) in the adipose tissues,and also upregulating glucagon-like peptide-1 (GLP-1) and serum insulin levels [36]. Simultaneously, the effects ofBacteroideson metabolic disorders caused by obesity were proved by facts that an increase in the relative abundance ofBacteroidesin the gut could reduce the risk of obesity [37,38].
3. Polysaccharide degradation by Bacteroides
3.1 Polysaccharide utilization loci and the Sus-like paradigm
Polysaccharides was the kind of carbohydrate with complex and huge molecular structure, which was formed by the condensation and dehydration of several monosaccharide molecules [39]. The capacity ofBacteroidesfor decomposing diet and mucosal polysaccharides,as well as those present on the surface of other gut microbes is the main factor that make them thrive in the environment of the gut. Up to now,studies have shown thatBacteroidescould utilize the polysaccharides in some similar ways (Fig. 1). The PULs were the gene clusters located in specific regions and encoded a series of enzymes for the capture,transport and degradation of polysaccharides [40]. Each PUL targeted a defined polysaccharide or a group of related polysaccharides [41].Genome sequencing and transcriptome data demonstrated that most of gutBacteroidesspecies had PULs, among whichB. thetaiotaomicronandB. ovatusdevoted up to 20% of their genomic content to these genetic clusters [24,42]. In addition,B. ovatuspossessed several PULs for the degradation of hemicelluloses, while the genome ofB. thetaiotaomicronhad a greater number of PULs involved in the metabolism of host mucin-O-glycans [43].B. fragiliscould efficiently harvest N-linked glycans from the glycoproteins common in the serum and serous fluid. This phenomenon was considered to be related to its 47 putative PULs and 22 stand-alone SusCD-like protein pairs [44]. Subsequent studies identified the existence and the functions of PULs in the genomes of human gut microbes, includingB. cellulosilyticusWH2,B. xylanisolvensXB1A andB. vulgatusATCC8482 [44-46].
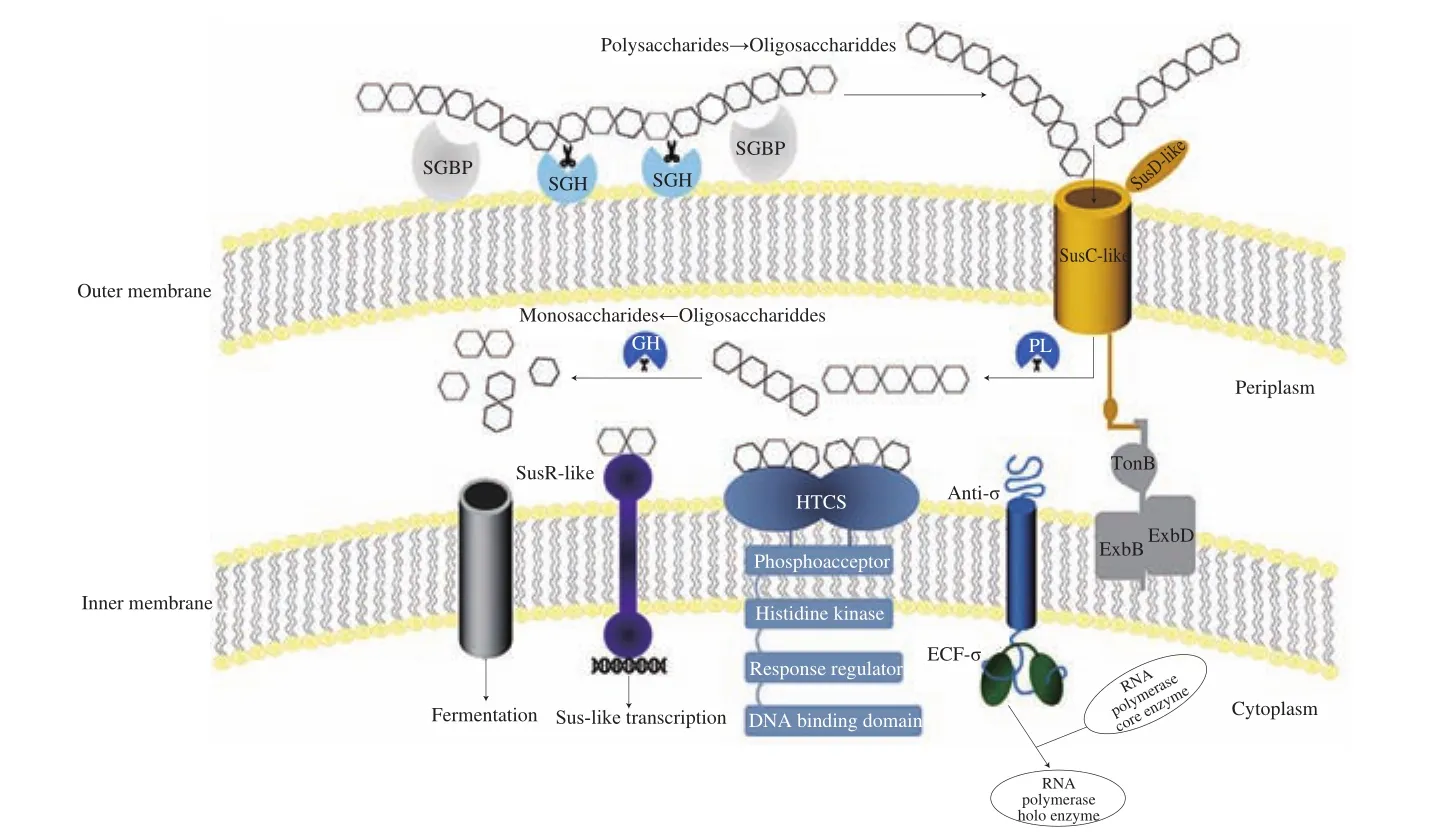
Fig. 1 The polysaccharide utilization system of Bacteroides. Polysaccharide was bound to the cell surface by the surface glycan-binding proteins (SGBPs).Subsequent hydrolysis by surface glycoside hydrolases (SGHs) generated the small oligosaccharides to transport through the SusC-like protein. The transport activity of the SusC-like protein was dependent on the energy provided by the TonB-ExbBD complex. Once in the periplasm, polysaccharide lyases (PLs) and glycoside hydrolases (GHs) processed oligosaccharides into smaller units or monosaccharides. Three inner membrane-spanning regulators (ECF-σ/anti-σ, HTCS,SusR like) sensed the existent of oligosaccharide degradation products and then drove the transcription of genes involved in the transport and degradation of the target polysaccharide. Disaccharides or monosaccharides were then shuttled into the cytoplasm for fermentation via a specific transporter.
PULs contained genes encoding three types of inner membrane sensor-regulator systems: SusR-like regulators, ECF-σ/anti-σ pairs,and HTCSs [47]. Generally, the activity of ECF-σ factors was inhibited by anti-σ factors. When the oligosaccharide fragments degraded from the polysaccharide were transported into the periplasm by a SusC-like transporter, the ECF-σ factor was released and interacted with core RNA polymerase to form the holo-enzyme,which resulted in the transcription of the target genes encoding the structure and enzyme components of the Sus-like system [23].Meanwhile, HTCSs combined all of the domains of classical two-component systems into a single protein that shared homologous periplasmic sensor domains belonging to the third most populated (HK3) family [48]. Members of the SusR family were predicted inner membrane-spanning receptors that included two parts, namely,periplasmic domains implicated in starch-derived oligosaccharides sensing and cytoplasmic domains triggering the upregulation ofsusgenes [49]. Most of the sensor domains contained a seven-bladedβ-propeller fold, which identified unique oligosaccharides with a length of 2-8 subunits [43,50]. Xylose, arabino-octaose and unsaturated chondroitin disaccharide activated corresponding PULs by directly binding to the respective HTCSs [45,51,52]. The regulation of PULs was commonly coordinated by ECF-σ/anti-σ pairs, which were associated with PULs targeting host-derived polysaccharides,or HTCSs associated with the PULs targeting various plant-cell-wall polysaccharides.
A series of membrane protein complexes encoded by PULs were responsible for the catabolism of many polysaccharides, which included the Sus-like systems derived from the starch utilization system inB. thetaiotaomicron[53]. Sus loci were made up of 8 adjacent genes,susRABCDEFG, including one regulatory gene,susR, and 7 structure genes [54]. SusDEFG contained a bacterial signal peptidase II recognition motif, which were membrane-attached lipoproteins. After the signal peptide sequence was excised under the action of signal peptidase, theN-acyl-S-diacylglyceryl moiety was covalently linked toN-terminal cysteines to form anN-terminalN-acyl-diacylglycerylcysteine through which lipoproteins were attached to the outer membrane [55]. SusDEFG possessed the starch-binding domains on the surface ofB. thetaiotaomicron,however, they played the distinct roles within the Sus [56,57]. SusC was a member of the TonB-dependent transport (TBDT) family which contained two conserved domains: a plug-like domain and a barrel-shaped domain composed of 22 antiparallelβ-strands which could transport rare nutrients and macromolecules via energy derived from the protonmotive force and the TonB-ExbBD complex [58,59].When maltose or starch-derived oligosaccharides were present, SusR could acted as a regulator to promote the expression of Sus structural genes [60]. The periplasmic SusA and SusB completely hydrolyzed theα-1,4 andα-1,6 linkages in starch oligosaccharides [61].Although PULs encode a number of polysaccharide-degrading enzymes, most of them were homologs of the Sus proteins. The ubiquity ofsusCD-like in PULs as well as the structure and sequence conservation of these proteins indicated the conserved mechanism for the polysaccharide import. Outer-membrane-tethered surface glycan-binding proteins (SGBPs), SusEF-like proteins, encoded by thesesusE-positioned genes (genes located downstream of thesesusD-like homologs) had no homology to SusEF, which appeared to show the similar functions in polysaccharide binding [62]. SGBPs bound a kind of special polysaccharide to the cell surface of the gut, and the polysaccharide was partially degraded extracellularly by a surface glycoside hydrolase (SGH) from the bacteria [63]. The released oligosaccharide products were then transported into the periplasm under the cooperation of SusCD-like proteins [64]. In the periplasm, glycoside hydrolases and polysaccharide lyases cleaved oligosaccharides into smaller units or monosaccharide to introduce them into cytoplasm via an unidentified transporter [65]. Sus-like paradigm encoded by thousands of PULsgenes present in sequencedBacteroidesemploys multiple cell membrane-associated proteins to catalyze polysaccharide degradation, while the type and quantity of these catalysts depend on the complexity of targeting substrates.For instance, due to the apparent diversity ofBacteroidesSus-like systems,B. thetaiotaomicronSus was the tip of the iceberg and the molecular strategy for starch utilization has been expanded to target other polysaccharides. It is the living way that enablesBacteroidesevolve to make the efficient use of a very broad suite of biofuel production.
3.2 Specificity of polysaccharide utilization in Bacteroides species
Bacteroidesspecies would differ in their PULs and the metabolic capacities specified by the genes in PULs which were activated in the face of some polysaccharides. For example,B. thetaiotaomicronencoded PULs for sulfatases which were critical to integrate host glycans such as heparin/heparan sulfate into glycolysis [66].Heparin (Hep) and heparan sulfate (HS) were linear polysaccharides comprising a disaccharide repeating unit of aD-glucosamine (GlcN)1,4 linked to a uronic acid (UA),D-glucuronic (GlcA), orL-iduronic acid (IdoA). The variable substitutions of disaccharide units, such asN-acetyl,O-sulfate andN-sulfate, constituted the complex sequences [67].Growth of theB. thetaiotaomicroncultured on HS was accompanied by the upregulation of PULHep, extending fromBt4652toBt4675,which was in response to this glycosaminoglycan (GAG) [40].Additionally, the sulfatase BT4656 and BT1596 were upregulated by the stimulation of Hep [68]. The surface lyase BT4662PL12was active on Hep,N-acetyl-de-O-sulfated Hep, and HS, with a preference for the latter two substrates. Furthermore, the depolymerization values were approximately 5%, 40% and 80%, respectively [69].The capacity of the polysaccharide lyases (PLs) and glycoside hydrolases (GHs) encoded by PULHepinB. thetaiotaomicronwere determined against the Hep-based GAGs [69]. Three periplasmic PLs,BT4652PL15, BT4657PL12, and BT4675PL13, resulted in the production of disaccharides which was a signaling molecule upregulating the PULHepduring the initial phase of degradation. BT4658GH88, which cleaved the glycosidic linkage between the unsaturated UA and GlcN/GlcNAc, only accommodated the sulfation at N2 or O6 of neutral sugar, reflecting the binding specificity of PULHepHTCS BT4663 [69,70].B. thetaiotaomicronmade the utilization of HS and Hep in a high priority position, whereasB. ovatuslacked the enzyme necessary for GAGs breakdown. Xylan-degradation system inB.ovatushad been characterized as a microbial resource allocation example. The organism, which could utilize the three major xylans,contained two discrete loci (PUL-XylS and PUL-XylL) encoding the xylan-degrading apparatus [71].
Bacteroidesspecies that utilized the glycan chondroitin sulfate (CS) had difference in the way of regulating orthologous CS utilization genes [72]. The Regulator BT3334 inB. thetaiotaomicronactivated by unsaturated disaccharides could control the transcription of GH-encoding genes which produced lyases cleaving CS in the periplasm. The sulfatase 1 (BT3333) mRNA, which represented the expression of BT3334-regulated genes, initially underwent a transcriptional surge when the inducing ligand for BT3334 was produced. Then the inducing ligand for BT3334 was dropped into the lower steady-state levels when the capacity of the rate-limiting glucuronyl hydrolase rose [51]. These findings revealed the mechanism thatB. thetaiotaomicrondynamically adjusted the transcription of CS utilization genes in response to the CS catabolic rate in the gut environment [72]. In contrast, the mRNA ofB. cellulosilyticussusC-like gene, which was the most induced transcription of the CS utilization genes, exhibited sustained upregulation upon being shifted into minimal medium containing CS [72]. Therefore, although the conserved sequences of PULs mediating degradation of special polysaccharides within these relatedBacteroidesstrains, ecological and other factors could still drive the difference among transcriptional behavior.
3.3 Competitive and symbiotic interactions between Bacteroides and other gut microbes
The Sus-like systems ofBacteroidesrepresented a solution for capturing carbon sources, which could avoid the competition with cellulosomes or extracellular freely diffusing enzymes employed by other bacteria. However, recent researches suggested that these apparently sel fish systems may share partial degradation products of polysaccharides with the nearby gut microbes [8]. Thus, the utilization of dietary polysaccharides byBacteroideswas of great significance for understanding how gut bacterial species interact with each other (Table 2).

Table 2Utilization of dietary polysaccharides by different Bacteroides species.
Bacteroidesspecies could degrade polysaccharides through a selfish mechanism to prevent other bacteria from utilizing their polysaccharide breakdown products (PBPs). For example, xylan degrading apparatus inB. ovatusseemed to be optimized to maximize the intracellular breakdown, which indicated that the organism adopted a selfish strategy while deconstructing the hemicellulose [71,73].B. thetaiotaomicronPULs (MAN-PUL1, MAN-PUL2 and MAN-PUL3)only exhibited the limited cleavage of bacteria cell wallα-mannan outside the bacteria cell and the primary hydrolysis ofα-1,6-mannose backbone subsequently happened in the periplasm [74]. Cellulosomes were multi-enzyme complexes formed by various cellulases and hemicellulases depending on anchoring-adhesion mechanism [75].In addition to PULs, cellulosomes mostly produced byClostridiumandRuminococcusrepresented another way of polysaccharide degradation [76]. Different from the whole binding and degradation of polysaccharides by cellulosomes on the surface of the bacterial cells, PULs only extracellularly depolymerized the polysaccharides into specific oligosaccharides, which could be recognized and utilized by limited number of bacteria. These phenomena were beneficial for making the bacteria encoding PULs obtain the dominance in the gut [7].
Symbiotic interactions betweenBacteroidesand other gut microbes were also found in the polysaccharide utilization. In co-culture fermentations, the proliferation ofBifidobacterium animalissubsp.lactiswas induced by xylooligosaccharides derived from the hydrolysis of xylan byBacteroidesspecies [77]. A model of gnotobiotic mice colonized withEubacterium rectaleand/orB. thetaiotaomicronprovided the evidence for the cross-feeding interactions between these two species [78].B. thetaiotaomicronadapted to the presence ofE. rectaleby upregulating the expression of genes located in PULs to degrade an increased variety of polysaccharide substrates and share the PBPs withE. rectale. In contrast, polysaccharide metabolism genes among theE. rectalegenes, particularly GHs, were downregulated in the presence ofB. thetaiotaomicron. Meanwhile, three strongly upregulated genes (EUBRE_3689,EUBREC_0479, andEUBREC_1075-6) were predicted to transport the cellobiose, galactoside, and arabinose,which suggested thatE. rectalewas more inclined to utilize the PBPs generated byB. thetaiotaomicron[78]. A fewBacteroidesspecies could release outer membrane vesicles (OMVs) which contained GHs needed to break down some polysaccharides [79,80].B. thetaiotaomicronreleased OMVs containing levanase responsible for the liberation of extracellular fructo-oligosaccharides which supported the growth of otherBacteroidesspecies. Similarly,B. ovatuscould secret the enzymes which were unnecessary for the utilization of inulin, but only to support the growth ofB. vulgatusin the inulin medium [17]. The OMVs-mediated polysaccharide utilization would change the formation of ecological networks among different bacterial species in the gut.
4. Bene ficial effects of Bacteroides-mediated polysaccharide metabolites on gut health
Bacteroidesdegradation on polysaccharides and cooperation with other gut microbes led to the formation of various products in the gut. Among them, SCFAs were the important metabolites. Acetate was produced via acetyl-CoA and Wood-Ljungdahl pathway from pyruvate [81,82]. Propionate could be generated by succinate and propanediol pathways, while the main sources of butyrate were from phosphotransbutyrylase/butyrate kinase and acetate CoA-transferase pathways [83]. However, what proportion of SCFAs contributed directly byBacteroidesin the gut had not been fully known, but they cleaved polysaccharides into oligosaccharides and monosaccharides to provide substrates for the fermentation of other SCFAs-producing bacteria, which revealed their involvement as mediators in the production of SCFAs. Fig. 2 showed how SCFAs affected the host gut health.
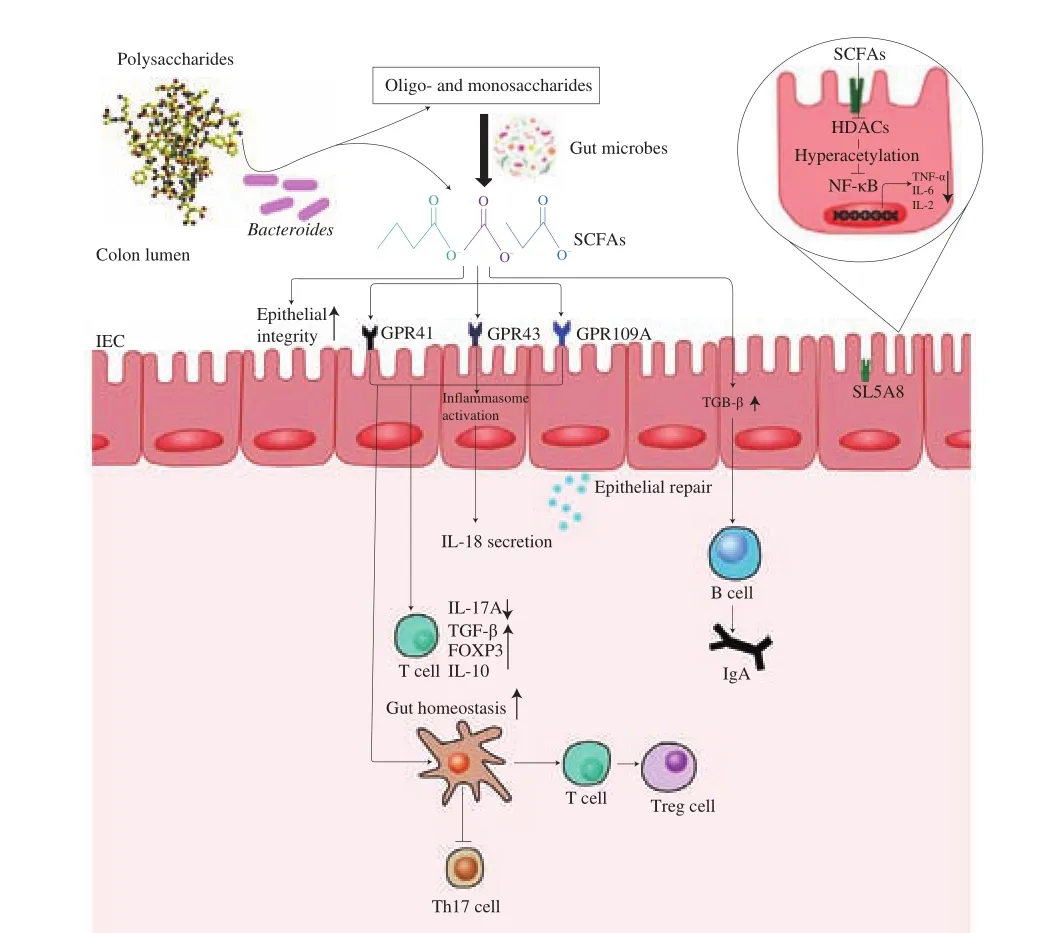
Fig. 2 The beneficial effects of SCFAs in intestinal inflammation and gut immunity.Bacteroides catabolized the polysaccharides into oligosaccharides or SCFAs.Fermentation of oligo- and monosaccharides by SCFAs-producing bacteria increased the luminal concentrations of acetate, butyrate, and propionate. In colonocytes, SCFAs inhibited histone deacetylases (HDACs) and NF-κB-induced proinflammatory cytokines expression. Lumina SCFAs sensed by GPR41, GPR43 and GPR109A activated the inflammasomes, which promoted the secretion of IL-18. SFCAs also served as the main energy source for colonocytes and induced the intestinal epithelial cells (IECs) to secrete antimicrobial peptides and mucins, thereby they could maintain the integrity of the epithelial barrier. In lamina propria, SCFAs reduced the inflammation by increasing the expression of anti-inflammatory factors and immunoglobulin A (IgA), as well as inducing the proliferation and the differentiation of the T cells.
SCFAs were closely related to the intestinal inflammation, which could be used as the signal molecules both inside and outside the bacteria cells to achieve anti-inflammatory effects. G protein-coupled receptors (GPCRs) were the largest family for the cell membrane surface receptors in the gut [84]. SCFAs were used as GPCRs agonist to extracellularly combine with three types of GPCRs, including GPR41, GPR43, and GPR109A. These receptors had different intracellular signaling capabilities. For example, GPR43 were reportedly to couple with either Gqor Gi/oand GPR41 exclusively activated Gi/opathway [85]. It was believed that the butyrate bound to GPR41 with high affinity, whereas acetate and propionate bound to GPR43 [86]. For instance, SCFAs binding to GPR43 and GPR109A on colonic epithelial cells stimulated K+ef flux and hyperpolarization,which led to NLRP3 inflammasome activation and increased the levels of interleukin (IL)-18 in serum, in a mouse model of DSS-induced colitis [87]. In addition, the administration of SCFAs was protective for colitis-associated colorectal cancer development by suppressing the expression of proinflammatory cytokines including IL-6, TNF-α and IL-17, which was also associated with a significant increase in the expression of GPR43 in colonic tissue [88]. Similarly, the butyrate displayed an anti-inflammatory effect via the inhibition of nuclear factor kappa B (NF-кB) activation [89]. Moreover, the production of IL-22 in CD4+T cells and innate lymphoid cells (ILCs) were upregulated by SCFAs through GPR41 [90]. SCFAs were transported into the bacteria cells through sodium-coupled monocarboxylate transporter 1 (i.e. SLC5A8) to regulate the inflammation as an inhibitor of histone deacetylases (HDACs) [91]. For example,butyrate exerted the anti-inflammatory effects on colonic lamina propria macrophages via the inhibition of HDACs. This process mainly included the downregulation of lipopolysaccharide-induced proinflammatory mediators, nitric oxide, IL-6, and IL-12 [92]. It was reported that the inhibition of HDACs in T cells induced by SCFAs could increase the acetylation of p70 S6 kinase and phosphorylation rS6, which regulated the mTOR pathway required for the generation of IL-10+T cells to alleviate the inflammation [93].
SCFAs played the key role in maintaining the intestinal immune homeostasis. Monocytes and neutrophils, as the natural immune cells, recognized and eliminated antigens to activate the adaptive immune response and prevent the pathogen invasion [94,95]. SCFAs could induce the neutrophil chemotaxis or regulate T cells (Treg cells) proliferation through binding to the GPR43 [96,97]. SCFAs also upregulated the expression ofFOXP3by inhibiting the HDAC, which then promoted the T cells to differentiate into the effector T cells and the Treg cells [93,98]. When IL-4-induced macrophages were co-cultured with the butyratein vitro, they displayed the enhanced phagocytosis and significantly suppressed the nitric oxide, IL-12p40 and IL-10 production [99]. In addition, butyrate and acetate could accelerate the secretion of IgA by increasing TGF-β in intestinal epithelial cells (IECs) or aldehyde dehydrogenase 1 family subfamily A2 (ALDH1A2)expression in dendritic cells (DCs), respectively [100,101].
5. Conclusions and perspectives
Research on the relationship between polysaccharide and gut microbiota had elucidated thatBacteroidespossessed the capacity to degrade various polysaccharides. This review systematically summarized theBacteroidesutilization on polysaccharides and their beneficial effects on the gut health, which revealed that it was necessary to analyze the whole metabolic products and keyBacteroidespecies for understanding the potential mechanism.
In future, researchers are suggested to improve the understanding of polysaccharide utilization from several aspects. Firstly, unrevealed mechanisms employed by gut microbiota for polysaccharides deserve more attention because some special species showed significant differences in the utilization of highly complex glycan, which may be species or even strain-dependent. Proteomics, metabolomics,and bioinformatics can be carried out to set up a comprehensive evaluation for the structure-activity relationship of polysaccharide during the degradation processesbyBacteroides. Secondly, the mechanisms of regulating systems of PULs that respond to distinct polysaccharides and their signal transduction pathways need further investigation. Thirdly, based on dynamic imaging and fluorescent probe labeling techniques, it is meaningful to visualize the real-time dynamics of uptake and utilization of polysaccharides byBacteroides.This could help us to find more subcellular localization of enzymes and understand how differentBacteroideswork together. Finally,polysaccharides or their corresponding metabolites resulted from the degradation byBacteroidescould be used as the therapeutic approaches for the diseases related to the gut dysbiosis.
Declaration of competing interest
All the authors declared no conflicts of interest.
Acknowledgments
The financial supports are coming from the National Science Fund for Distinguished Young Scholars of China (31825020).
- 食品科学与人类健康(英文)的其它文章
- Hypoglycemic natural products with in vivo activities and their mechanisms: a review
- Capsular polysaccarides of probiotics and their immunomodulatory roles
- Natural compounds may contribute in preventing SARS-CoV-2 infection: a narrative review
- A comprehensive review on the effects of green tea and its components on the immune function
- A review on current and future advancements for commercialized microalgae species
- Effects of silkworm pupa protein on apoptosis and energy metabolism in human colon cancer DLD-1 cells

