Force-constant-decayed anisotropic network model: An improved method for predicting RNA flexibility
Wei-Bu Wang(王韦卜) Xing-Yuan Li(李兴元) and Ji-Guo Su(苏计国)
1Key Laboratory for Microstructural Material Physics of Hebei Province,School of Science,Yanshan University,Qinhuangdao 066004,China
2The Sixth Laboratory,National Vaccine and Serum Institute,Beijing 101111,China
Keywords: anisotropic network model,B-factor,RNA
1. Introduction
RNA plays a critical role in the maintenance of cell activities, such as gene delivery, translation and splicing.[1–5]For a long time, RNA was known as the information transmitter between protein and DNA in life processes, which was considered to be only a single-strand chain without fixed three-dimensional structure. However, in recent years several kinds of RNA, such as riboswitches, ribozymes and protein-complexed RNAs,have been proved to be folded into sophisticated tertiary structures just like proteins, and undergo subtle conformational changes to perform their unique functions.[6–10]The single-molecule experimental results also indicated that even in small RNA molecules, the structural dynamics could be remarkably complex and intricately coupled with special functions.[11]Therefore, extracting dynamical properties from RNA tertiary structure is important not only for understanding RNA functions, but also for developing novel RNA-targeted or RNA-based therapeutics.[12,13]
Molecular dynamics (MD) simulation is a common method to investigate dynamical properties of biomacromolecule,which has been widely used to explore protein flexibility and its relationship to protein function in atomic levels for over 40 years.[14]With the development of the force field,MD simulation has also been extended to study the relationship between the structural flexibility and biological function of RNAs.[15–17]Drozdetskiet al. investigated the influence of multivalent counterions on the flexibility of RNA by using MD simulations, and found that the counterions can significantly increase RNA bending persistence length.[18]Baoet al.explored the different flexibility between double-stranded RNA and DNA in the aspects of stretching and twist-stretch coupling by using MD simulations,and they revealed that the flexibility differences between these two structures were mainly attributed to different helical structures.[19]Deng and Cieplak used MD simulations to describe the folding progresses of two RNA hairpin structures qualitatively,and the potential of mean force was calculated to prove the stability of the folding structure.[20]However,MD simulation requires significant computer resources, and costs a lot of time to describe the large-scale conformational motions.[21]It is still not a trivial task to investigate the intrinsic large-scale dynamics encoded in RNA structures by using MD simulations.
Besides the MD simulation,elastic network mode(ENM)is another effective method to explore the conformational dynamics of biomacromolecules,which has been widely used in proteins.[22–26]ENM simplifies a biomacromolecule structure as a network of nodes,in which the interactions between nodes are described as harmonic springs with an uniform force constant. In ENM, the functional motions of the system can be captured by normal mode analysis. Many studies have proved that ENM is an efficient method to investigate the large-scale conformational movements in proteins, and the residue fluctuations calculated by the ENM are in good agreement with the temperature factors(B-factors)measured by x-ray crystallographic experiments.[27–29]ENM is divided into two categories, i.e., Gaussian network model (GNM) and anisotropic network model (ANM).[30,31]GNM assumes that the magnitudes of node fluctuations follow the Gaussian distribution around the equilibrium position, and the direction of fluctuations is not taken into account in the model. ANM can be considered as an extension of GNM, in which the directions of individual node fluctuations in the network are regarded to be anisotropic.[31,32]ANM has also been extensively used to describe the anisotropic conformational motions related to the biological functions of proteins.[33–35]In addition, ANM has also been extended to investigate the dynamics of RNAs and DNAs. Zimmermann and Jernigan have shown that ANM can capture the conformational motions observed in a collection of RNA ensembles of experimentally determined structures.[36]Wanget al.used the ANM to explore the modes of motions in the whole 70s ribosome,and they found that the slow modes correspond closely to the ratchet-like motion observed in experiments.[37]Wanget al.further proposed a new method based on ANM to investigate the conformational motions of subunits in large biological assemblies,in which the rigid body motions could be separated from the nonrigid body motions of the subunits.[38]Gonzalezet al.applied ANM to explore the conformational space of CUG repeats, and the result showed that ANM can extract global dynamics encoded in the CUG repeats structure.[39]
Although conventional ANM has been used in predicting the nucleotide anisotropic fluctuations of RNAs, the performance of ANM in predicting RNA flexibility is much poorer than that in proteins.[36,37]In the conventional ANM,a given cutoff distance is used to determine whether interactions exist between two nucleotides,and the inter-nucleotide interactions are simplified as springs with identical force constants. However,the nucleotides in RNAs are negatively charged,and the conventional construction of ANM based on a cutoff distance may result in the loss of the weak yet important long-range electrostatic interactions in the model. The cutoff-based ANM construction method may not be suitable for RNA systems.
In this work,a new model,named force-constant-decayed ANM(fcd-ANM),was proposed to better predict the dynamical properties encoded in RNA structures. In our method,all pairwise nucleotides were connected by springs,and the force constants of springs were decayed exponentially based on the separation distance to describe different interaction strengths between nucleotides. The force-constant-decayed method has been successfully used in GNM in our previous studies,[40]and in the present study this method was further extended combing with ANM to better describe RNA dynamics. The performance of the proposed fcd-ANM in predicting RNA flexibility was evaluated by using a non-redundant RNA structure database, in which theB-factors of nucleotides in these RNA structures were calculated by fcd-ANM and compared with the experimental data. Our calculation results show that fcd-ANM significantly outperforms the conventional ANM in predicting the experimentalB-factors of RNA structures,which may serve as a better model for RNA flexibility studies
2. Method
2.1. Conventional anisotropic network model
In the conventional anisotropic network model (ANM),RNA structures downloaded from Protein Data Bank (PDB)are simplified as elastic networks, in which the P atom of each nucleotide in RNA is represent as a node and all pairwise nucleotides within a given cutoff distance are connect by springs.[37,41]ANM contains two adjustable parameters: the cutoff distanceRcand the force constant of springsγ. In the conventional ANM,Rcis usually set to 20 °A–24 °A[20,38,42]andγis assigned a uniform value. All the springs in ANM are assumed to be in equilibrium in the initial structure, and the potential energy of the RNA system withNnucleotides is the sum of the harmonic energy of all the springs, which can be expressed as
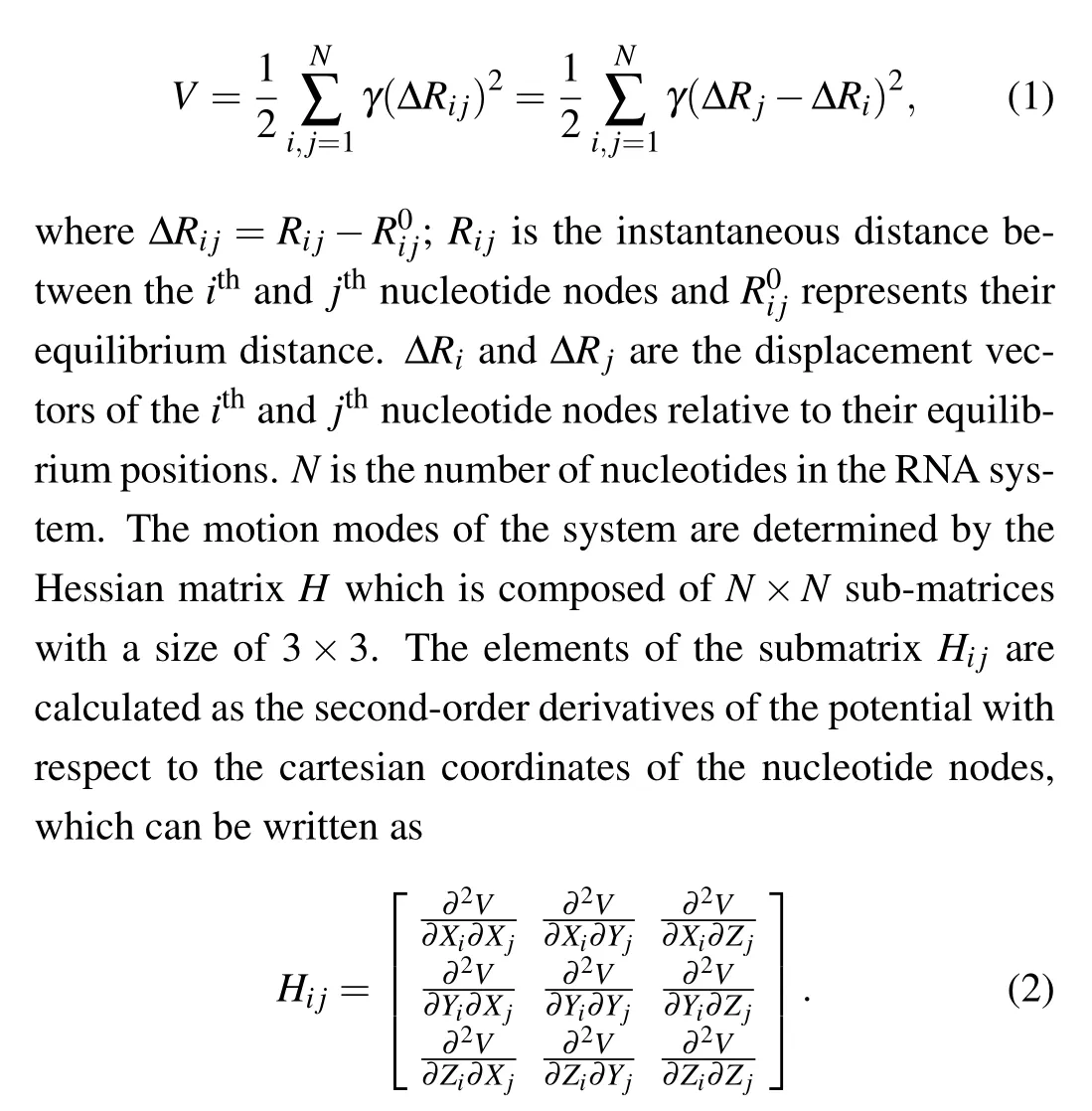
Based on the normal mode analysis theory, the normal modes of RNA can be calculated by the diagonalization of Hessian matrixH.The diagonalization ofHgenerates six zero eigenvalues and 3N-6 non-zero eigenvalues. The six zero eigenvalues correspond to the overall translational and rotational degrees of freedom of the system,which are not related to the internal motions within the RNA molecule. The dynamical properties of the system are determined by the 3N-6 normal modes with non-zero eigenvalues. Therefore, the meansquare fluctuation for each nucleotide node can be computed as
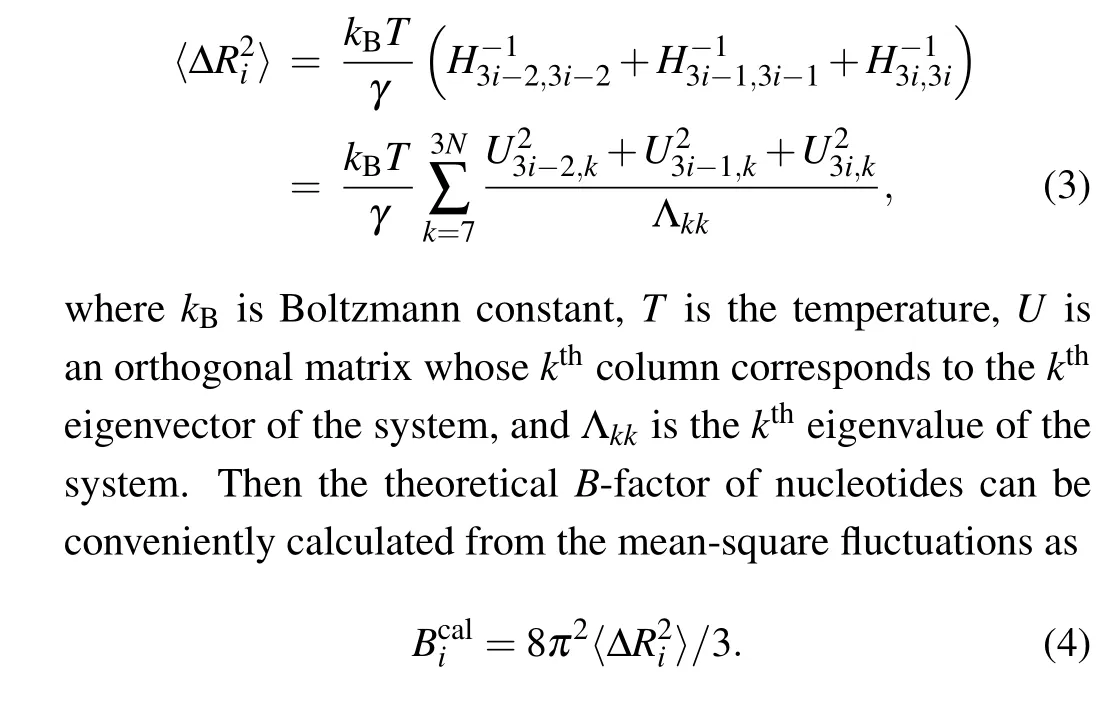
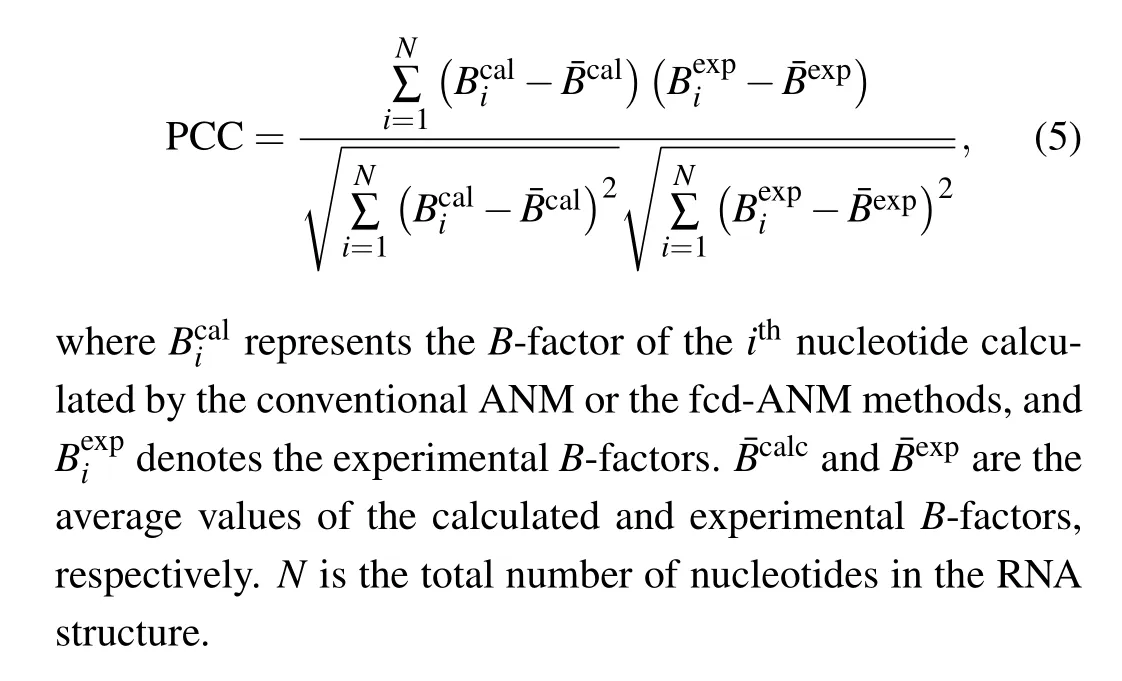
In this study, Pearson correlation coefficient (PCC) between the calculatedB-factors and the experimental data acquired from x-ray crystallographic method was computed to evaluate the performance of the conventional ANM as well as the fcd-ANM model. The PCC between the calculated and experimentalB-factors can be expressed as
2.2. Force-constant-decayed anisotropic network model
In the conventional ANM, only the pairwise nucleotides whose distance is less than the cutoff value is considered to be interacted with each other, and the force constants for all the inter-nucleotide interactions are identical. However,RNA structures contain various inter-nucleotide interactions with different strengths,in which the short-range covalent as well as base-base paring and stacking interactions are much stronger than the long-range electrostatic interactions. The uniform force constant in the conventional ANM cannot distinguish the differences in the inter-nucleotide interactions. Besides that,conventional construction of ANM based on a cutoff distance may result in the neglect of long-range electrostatic interactions that are also important in controlling RNA dynamics.Therefore, the conventional ANM construction method may not be suitable for RNA structures. In the present work, to better describe RNA structural dynamics, a new model, i.e.,fcd-ANM was proposed, in which all nucleotide pairs were connected by springs and the force constants of springs were decayed with the separation distance to consider different interaction strengths between nucleotides.
Similar to the conventional ANM,the RNA structure was modeled as an elastic network, in which each nucleotide was simplified as a node located on its P atom. But different from the conventional ANM, all pairwise nucleotides in the structure were connected by springs,and the spring force constants were decayed exponentially with their separation distance between pairwise nucleotides in fcd-ANM,which can be written as
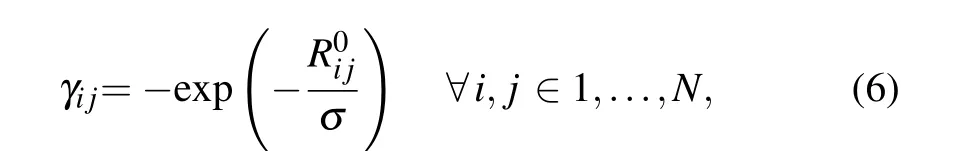
whereR0ijrepresents the equilibrium distance between theithandjthnucleotides;Nis the total number of the nucleotides in RNA;σis a variable parameter, which controls the decay rate of the spring force constant with changes of the separation distance between nucleotides. Based this new model,the potential energy and Hessian matrix for fcd-ANM can be calculated according to Eqs. (1) and (2). Then, theB-factors of nucleotides can be calculated by using Eqs. (3) and (4), and the PCC values between the computedB-factors and the experimental data derived from x-ray crystallography can be calculated by using Eq. (5). The PCC values obtained by the proposed fcd-ANM were compared with those by the conventional ANM to assess the performance of our model in predicting RNA flexibility.
2.3. Database of non-redundant RNA tertiary structures
In this study, the non-redundant RNA structure dataset constructed in our previous work[40]was used to test the performance of the proposed fcd-ANM model in predicting the flexibility of RNAs. The non-redundant RNA structures in the dataset were obtained from the representative set 3.128 of RNA 3D structures collected by Leontis and Zirbel.[43]The RNA structures meeting the following screening criteria were selected: (a) structures determined by x-ray crystallography;(b) structural resolution better than 3.0 °A; (c) the number of nucleotides larger than 122;(d)nucleotideB-factors available.Finally, 51 RNA structures were screened and included in the non-redundant RNA structure dataset,and the PDB codes of the filtered RNA structures can be found in our previous work,[40]which were also listed in Table S1 in the supplementary materials.
3. Result and discussion
3.1. Optimization of the parameters in the conventional ANM and the proposed fcd-ANM
The conventional ANM contains two variable parameters,i.e.,the force constant of springsγand the cutoff distanceRc.Because the value ofγhas no effect on the relative fluctuations of nucleotide, only the value ofRcwas optimized in this study.Rcvalue was changed from 17 °A to 27 °A with an interval of 1 °A, and the PCC between the predicted and experimentalB-factors was calculated at different values ofRcfor each RNA structure in the non-redundant database. The calculation results are given in Table S2 in the supplementary materials. The average PCC as a function ofRcis shown in Fig. 1(a). It is found that the average PCC is in the range of 0.309 to 0.437,and the value ofRchas obvious effects on the value of PCC. The peak value of the average PCC is 0.437,which corresponds to the cutoff distance of 21 °A.Hence, theRcvalue of 21 °A was used for the conventional ANM in this study.
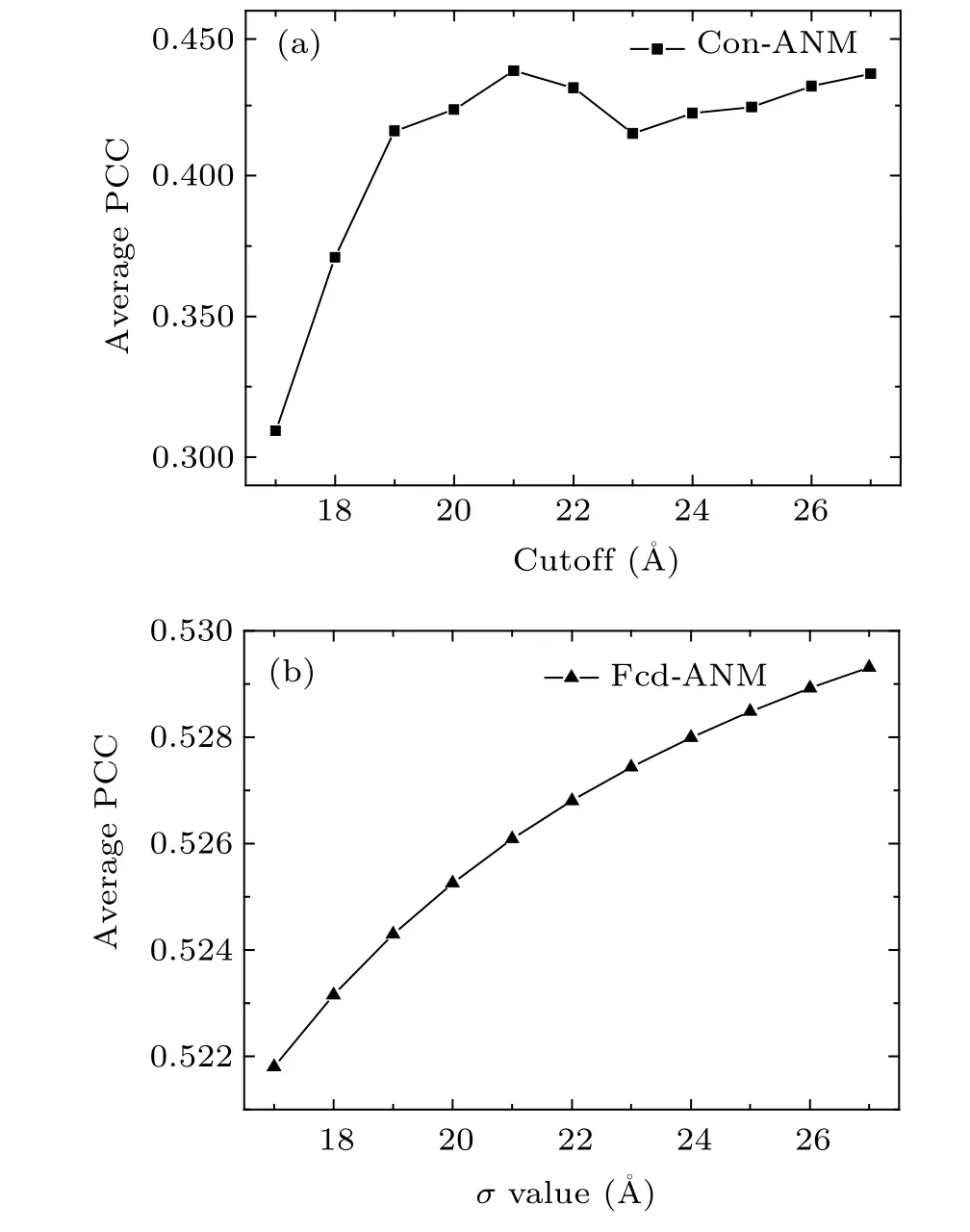
In the proposed fcd-ANM,σis the only adjustable parameter,which controls the decay rate of the spring force constant along with the increasing of the distance between pairwise nucleotides. In order to obtain the optimal value,σwas changed from 17 °A to 27 °A with the step of 1 °A,and the PCC values between the predicted and experimentalB-factors for all the RNA structures in the dataset were calculated at differentσvalues (Table S3 in the supplementary materials). The change of the average PCC with differentσvalues is displayed in Fig.1(b). The peak value of average PCC is 0.529 and correspondingly the optimalσvalue is determined to be 27 °A.Consequently,in this study 27 °A was adopted for the parameterσin fcd-ANM.
3.2. Improved performance of the proposed fcd-ANM in predicting RNA B-factors compared to the conventional ANM
A total 51 RNA structures in the non-redundant database were used to evaluate the accuracy of fcd-ANM in predicting the nucleotideB-factors of the systems, and the prediction accuracy of fcd-ANM was also compared with that of the conventional ANM. For each RNA structure, the nucleotideB-factors were predicted by fcd-ANM and the conventional ANM, respectively, in which the optimized values were employed for the adjustable parameters in the models as discussed in the previous section. Then, the PCC between the predicted and experimentalB-factors was calculated both for ANM and fcd-ANM. The PCC values for each of the RNA structures obtained by the conventional ANM and fcd-ANM are displayed in Fig. 2(a) and Table 1, and the comparation of the PCC values between these two models for all the studied RNA structures is shown in Fig. 2(b). There are 6 RNA structures whose nucleotideB-factors cannot be calculated using the conventional ANM,because their structures are loosely packed and more than 6 modes with zero frequency were obtained. The PCC values of these structures for the conventional ANM are set to zero in Fig. 2(b). The results show that for most of the RNA structures,the PCC values predicted by fcd-ANM are significantly higher than those by the conventional ANM. The average PCC for all the studied RNA structures increases from 0.437 to 0.529 for fcd-ANM in comparison to the conventional ANM.The prediction accuracy of fcd-ANM on RNAB-factors was significantly improved by 21.05%. These results indicate that fcd-ANM is superior over the conventional ANM in predicting nucleotide fluctuations in RNA tertiary structures.
As shown in Table 1, there are different types of RNA structures contained in the non-redundant database,including ribosomal RNA, riboswitch RNA, ribozyme RNA and other types. The average PCC values for these different types of RNA structure were also calculated separately to verify the effectiveness of fcd-ANM on different RNA types. For ribosomal RNA,the average PCC increased from 0.485 to 0.627,and thus the prediction accuracy was improved by 29.1%. For riboswitch RNA, the average PCC increased from 0.451 to 0.489, with a growth rate of 8.42%. For ribozyme RNA, the average PCC increased from 0.373 to 0.519, and the growth rate was 39.14%.These results indicate that fcd-ANM can significantly improve the prediction accuracy of nucleotide fluctuations for all these different RNA types.

As discussed above,for some loosely-packed RNA structures, theB-factors cannot be calculated using the conventional cutoff-based ANM,because the number of springs connected to some nodes in the network is too small. This problem can be solved in fcd-ANM.In this new model,all the pairwise nucleotides were connected by springs and the different interaction strengths between nucleotides were described by different force constants. The results in Table 1 show that theB-factors of the 6 RNA structures,unable to be computed by the conventional ANM,can be calculated using fcd-ANM.Therefore, fcd-ANM not only outperforms the conventional ANM in predicting RNA flexibility,but also can be applied to loosely packed RNA structures.

PDB code Chain Category ANM Fcd-ANM 6QZP L5+L8 ribosomal RNA 0.316 0.687 6SGC 54+84 ribosomal RNA 0.405 0.771 5TBW 1+4 ribosomal RNA 0.543 0.707 6S0Z A ribosomal RNA 0.762 0.774 5J7L DA ribosomal RNA 0.743 0.809 6SPB A ribosomal RNA 0.594 0.751 6V3A AN1 ribosomal RNA 0.600 0.724 4Y4O 2A ribosomal RNA 0.688 0.806 4V9F 0 ribosomal RNA 0.717 0.718 5DM6 X ribosomal RNA 0.663 0.744 6ERI AA ribosomal RNA 0.058 0.769 4V88 A6 ribosomal RNA 0.649 0.665 6AZ1 1 ribosomal RNA /a) 0.761 6QZP S2 ribosomal RNA 0.523 0.704 6SGC A1 ribosomal RNA 0.701 0.723 6AZ3 1+7 ribosomal RNA 0.469 0.571 6S0X a ribosomal RNA 0.325 0.448 5J7L AA ribosomal RNA 0.723 0.767 6SPC a ribosomal RNA 0.485 0.524 6V3A sN1 ribosomal RNA 0.573 0.576 4Y4O 1a ribosomal RNA 0.617 0.677 6TMF A ribosomal RNA 0.523 0.545 6ERI BA ribosomal RNA 0.084 0.764 5T5H A+C ribosomal RNA 0.351 0.391 5T2A B ribosomal RNA 0.088 -0.178 6AZ3 2 ribosomal RNA 0.116 0.787 5T5H B ribosomal RNA / 0.378 4FAR A ribozyme RNA 0.673 0.613 4P95 A ribozyme RNA 0.463 0.503 3JCS 3 ribosomal RNA 0.173 0.261 6AZ3 4 ribosomal RNA 0.731 0.615 6AZ3 3 ribosomal RNA 0.372 0.576 3DIL A riboswitch RNA 0.396 0.457 3D0U A riboswitch RNA 0.215 0.174 3PDR X riboswitch RNA 0.342 0.253 6UFG A riboswitch RNA 0.498 0.637 2R8S R ribozyme RNA 0.445 0.778 1U9S A ribozyme RNA 0.343 0.370 4ZDO E tRNA -0.029 -0.059 5T5H E srRNA 0.026 0.050 3G9C Q ribozyme RNA 0.100 0.361 6ICZ H+F snRNA / 0.264 3NDB M scRNA 0.422 0.404 3HHN E ribozyme RNA 0.133 0.381 6ID1 H+F snRNA / 0.204 6KC8 B sgRNA / 0.349 3V7E C riboswitch RNA 0.624 0.722 6N5P A riboswitch RNA 0.633 0.690 6QW6 4 snRNA / 0.123 1L9A B scRNA 0.348 0.279 2Z75 B ribozyme RNA 0.456 0.628 Average PCC 0.437 0.529
3.3. Case study for comparison of the performance between ANM and fcd-ANM in predicting RNA flexibility
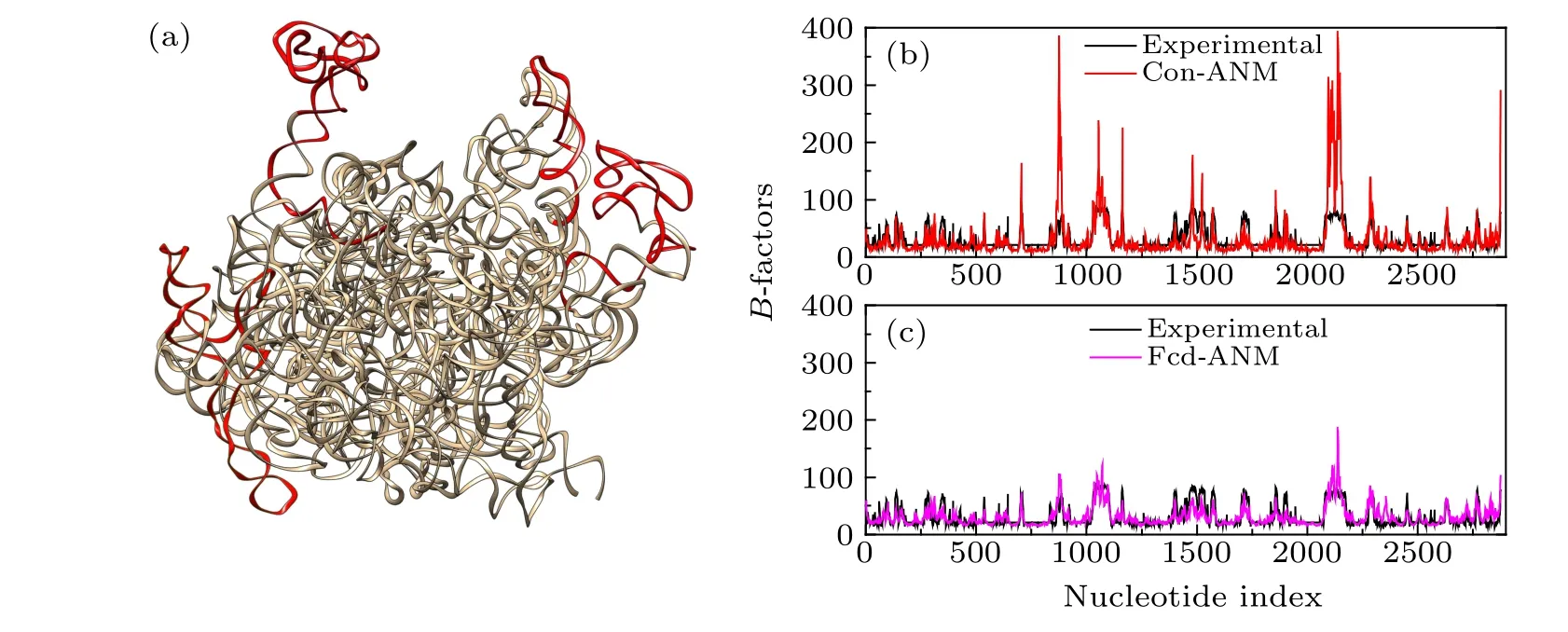
A RNA structure, i.e. the “AN1” chain of the acinetobacter baumannii ribosome (PDB code:6V3A),[44]was used as a case study to illustrate the better performance of fcd-ANM than that of the conventional ANM in predicting the nucleotideB-factors of the structure. The AN1 chain of the acinetobacter baumannii ribosome is a large RNA with 2877 nucleotides,which is composed of a well-packed core domain with some stems protruding out of the core structure,as shown in Fig.3(a). The nucleotideB-factors of the system were calculated by the conventional ANM and fcd-ANM respectively,and the PCC values between the calculated and experimentalB-factors were also computed for these two models. The PCC value for the conventional ANM was 0.600, whereas that for fcd-ANM was significantly improved to 0.724. The result indicates that fcd-ANM can better extract the flexibility encoded in the RNA structure than the conventional ANM.
TheB-factor profile of the studied RNA structure calculated by fcd-ANM was compared with that computed by the conventional ANM,as shown in Figs.3(b)and 3(c).It is found that theB-factors of nucleotides calculated by the conventional ANM were much higher than the experimental results in several peaks of the curve displayed in Fig. 3(b). These peaks correspond to the nucleotides 860-900,1022-1096,1475-1580 and 2072-2179,which protrude out from the core structure as highlighted by red in Fig.3(a). This result implies that in the conventional ANM,the cutoff-based construction of the model may be insufficient to restrict the flexible dynamics of these isolated substructures. By contrast,theB-factors predicted by fcd-ANM fit well with the experimental results, as shown in Fig. 3(c). Especially, for the protruding substructures corresponding to the peaks of theB-factor profile, the fluctuations predicted by fcd-ANM were obviously lower than those predicted by the conventional ANM, and were consistent better with the experimental data.All these results indicate that compared with the conventional ANM, fcd-ANM can better capture the inter-nucleotide interactions with different strengths in RNA structures and is more effective in exploring the intrinsic dynamics encoded in the structures.
4. Conclusion
Increasing evidence has demonstrated that RNA performs different kinds of functions in many biomolecular processes,and the functions of RNA are largely determined by its specific structure as well as the dynamics encoded in the structure. How to effectively extract the structure-encoded dynamical properties is of great significance for our understanding of the molecular mechanism behinds the RNA functions. The conventional ANM has been proved to be an effective method in investigating protein dynamical properties,however,the application of this method on the studies of RNA dynamics is not as efficient as that on proteins because of the different structural characteristics of RNAs in comparison to proteins.
In the conventional ANM, a cut-off value was used to determine the existence of interactions between pairwise nucleotides and a uniform force constant was applied for all the inter-nucleotide interactions. However, the tertiary structure of RNAs is usually loosely packed and there exist many longrange interactions between nucleotides in RNA structures.The cutoff-based construction method in the conventional ANM is not suitable for RNA structures. Therefore, in this study we proposed a new approach, named fcd-ANM, to improve the performance of ANM in investigating the structural dynamics of RNAs. In fcd-ANM, all nucleotide pairs in the RNA structure were connected by springs, and the force constants of springs were decayed exponentially as a function of the distance between nucleotides. The effectiveness of fcd-ANM in predicting RNA flexibility was tested on a non-redundant database containing 51 RNA structures. The study results show that fcd-ANM is obviously superior to the conventional ANM in extracting the intrinsic dynamical properties encoded in RNA structures. Comparing with the conventional ANM,the average PCC value between the calculated and experimentalB-factors was substantially improved by 21.05% for fcd-ANM,indicating that the proposed model can better reproduce the experimentalB-factors. In addition, fcd-ANM can be applied to the extremely loose RNA structures whoseB-factors cannot be calculated by the conventional ANM.Our study provided an effective method for investigating RNA flexibility. It should be pointed out that although our model has been successfully applied in extracting the intrinsic flexibility of largesize RNAs, it is still a challenge to predict the structural dynamics of small-size RNAs. Compared with large-size RNAs,the small-size ones are more loosely packed and usually interact with other macromolecules, such as proteins, to stabilize their structures. For investigation of the dynamics of smallsize RNAs, the interacted proteins should be considered into the model. Besides that,more intricate models,in which each nucleotide is simplified,for example,as three nodes instead of only one node, should be constructed to better exploring the flexibility of small RNAs.
ENM has also been employed by many groups to investigate the intrinsic flexibility of DNAs,[45–48]and it has been proved that ENM can capture many dynamical properties encoded in the DNA structure. Our model is developed based on ENM,and thus it can also be used in DNA flexibility studies. However, the investigation of subtle dynamics encoded in RNA and DNA structures may need more dedicated ENM models that take more structural details into account. We will improve the model to consider more structure details in the future.
Data availability
The data that support the findings of this study are openly available in Science Data Bank at http://doi.org/10.57760/sciencedb.j00113.00029.
- Chinese Physics B的其它文章
- Ergodic stationary distribution of a stochastic rumor propagation model with general incidence function
- Most probable transition paths in eutrophicated lake ecosystem under Gaussian white noise and periodic force
- Local sum uncertainty relations for angular momentum operators of bipartite permutation symmetric systems
- Quantum algorithm for neighborhood preserving embedding
- Vortex chains induced by anisotropic spin–orbit coupling and magnetic field in spin-2 Bose–Einstein condensates
- Short-wave infrared continuous-variable quantum key distribution over satellite-to-submarine channels