Mapping big brains at subcellular resolution in the era of big data in zoology
Connectome-scale structural mapping is fundamental for understanding the underlying mechanisms of brain cognition and brain disease pathogenesis.By combining rapidly developing three-dimensional (3D) imaging techniques and big data analysis methods,researchers are working on mesoscale mapping of mammalian brains at an accelerated pace.Here,we briefly describe existing brain-wide imaging strategies,especially our recently established primateoptimized pipeline capable of pan-brain neuronal connectivity mapping at subcellular resolution,and further discuss their vast application prospects in the big data era of zoology.
As interdisciplinary technologies and “omics” analyses revolutionize many biological domains,connectomics research,i.e.,the study of connectivity or “wiring” at different scales in the brain,has become a rapidly moving frontier.In particular,at the mesoscopic level,generating 3D cellular connectivity maps of the mammalian brain has been an overarching goal of major brain research initiatives in many countries (Poo et al.,2016).Because axons of individual neurons can target cells in multiple brain areas far from their soma,brain-wide maps of neuronal projections at subcellular or axonal resolution are essential.Such mapping efforts will establish a foundation for understanding the principles of brain wiring and information flow,and thus the mechanisms underlying specific brain functions and diseases.
Mesoscopic mapping of neuronal projections in the rodent brain has progressed considerably in the past decade (Gao et al.,2022;Peng et al.,2021;Winnubst et al.,2019),largely thanks to advances in imaging techniques such as fluorescence micro-optical sectioning tomography and serial two-photon tomography.With these techniques,the entire mouse brain can be imaged within several days.Another approach based on whole-brain tissue clearing and light-sheet microscopy (Ueda et al.,2020) can achieve high-speed imaging of the mouse brain at cellular resolution within hours.However,due to the large size and intrinsic inhomogeneity of brain tissue,this approach cannot achieve the subcellular resolution uniformity necessary for brain-wide axonal tracking.
For larger mammals,such as non-human primates,panbrain connectome mapping is a major challenge.Compared to mouse brains,macaque brains are over 200-fold larger and require much higher speeds for 3D imaging at sufficient resolution.Furthermore,once monkey brain imaging is complete,the resulting petabytes of data require efficient algorithms to reconstruct accurate 3D structures and analyze neuronal fibers and connectivity.
In a recent paper in Nature Biotechnology (Xu et al.,2021),we reported a systematic strategy to overcome the above bottlenecks (Figure 1) based on the Volumetric Imaging with Synchronized on-the-fly-scan and Readout (VISoR) approach.This scalable method enables ultra-high-speed 3D imaging of cleared brain slices with the continuous capture of blur-free,submicron-resolution images of moving samples (Figure 1A)(Wang et al.,2019).We improved the VISoR system and established a primate-optimized pipeline,i.e.,serial Sectioning and clearing,3D Microscopy with semi-Automated Reconstruction and Tracing (SMART) (Figure 1B) (Xu et al.,2021),enabling high-throughput connectome-scale mapping of macaque brains at micron resolution.With this approach,we first sectioned a whole rhesus macaque brain into consecutive coronal slices (300 μm thick),then cleared each slice independently with primate-optimized membrane permeabilization and high-refractive index matching solutions,allowing uniform transparency through both gray and white matter.With the improved VISoR system,we completed image acquisition of all cleared slices of the entire macaque brain within 100 hours.Notably,a new approach combining line-scan microscopy with integrated vibratome was recently developed for 3D imaging of large embedded samples at subcellular resolution (Zhou et al.,2022),which was capable of imaging an entire macaque brain within 70–80 days.
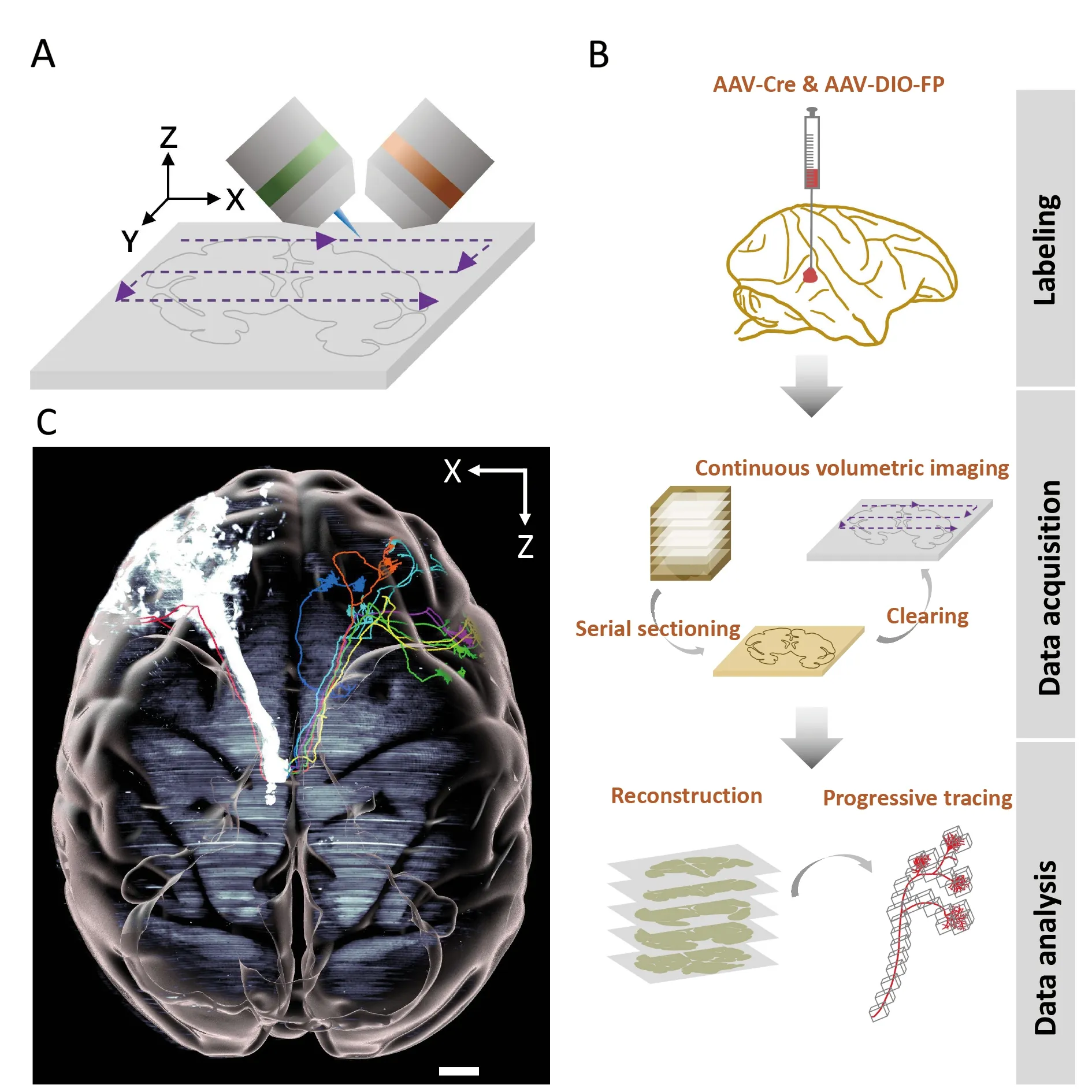
Figure 1 Mapping axonal projections in a rhesus macaque brain with VISoR imaging and SMART pipeline
For the VISoR/SMART approach,we developed automated reconstruction software to stitch image sets from nonoverlapping adjacent slices.We also developed semiautomated software capable of efficient progressive fiber tracking through the huge dataset of monkey brain images(Figure 1B).Using VISoR imaging and the SMART pipeline,we performed mesoscopic mapping of thalamocortical projections in a rhesus macaque brain (Figure 1C) (Xu et al.,2021),showcasing its potential capacity to construct detailed connectivity maps,especially when combined with sparse labeling strategies.In addition,we performed brain-wide single-fiber tracking based on the micron-resolution imaging accuracy and semi-automated annotation tools.Our preliminary observations indicated that thalamic neuronal axons have diverse projection patterns with unexpectedly complex routes,especially within cortical folding areas.Along these routes,we observed that some fibers turned sharply in white matter,which may be related to cortical folding (Xu et al.,2021).These findings demonstrate the important capability of whole-brain structural analysis at subcellular resolution,especially for understanding brain morphogenesis and neural circuit establishment (Van Essen,2020).
Given the unique role of non-human primates as animal models for studies on human cognitive functions and diseases,it has become increasingly important to analyze their brain structures at different scales (Chen et al.,2020;Xu et al.,2021;Xu et al.,2022;Zhou et al.,2022).The highthroughput VISoR/SMART system enables structural studies of the primate brain at unprecedented scale and detail,e.g.,large-scale pan-brain characterization of neuronal morphology,an essential step for understanding the principles of brain wiring diagrams.VISoR imaging can also be combined with other omics techniques to generate comprehensive brain maps,such as spatially defined gene expression atlases.Together with mesoscopic connectome mapping,these atlases could help reveal the neuroanatomic mechanisms underlying complex brain functions from a more holistic perspective,especially when complemented by functional approaches such as electrical microstimulation and functional magnetic resonance imaging (fMRI) (Xu et al.,2022).Furthermore,with recent advances in gene-editing in non-human primate brain disease models (Zhou et al.,2019),high-throughput connectome imaging could help unravel the complex circuit mechanisms of brain disorders in humans.
Beyond the brain,VISoR imaging can also be adapted to study diverse organs,even whole bodies of small and large animals,and thus promises to be useful tool for other zoological research such as animal development and systems physiology.As such,VISoR and other high-throughput microscopy methods are expected to generate an enormous amount of data across entire organs and systems with unprecedented cellular detail,which will undoubtedly lead to novel and groundbreaking discoveries.Furthermore,highresolution imaging of large samples will also generate huge datasets.Extracting valuable information from these massive datasets will require efficient algorithms,including AI-based image processing,other systematic strategies such as crowdsourcing and manual computation (Gao et al.,2022),as well as dedicated high-performance computing resources.Going forward,this interdisciplinary synergy in the era of big data not only heralds potential new discoveries,but also a possible paradigm shift in zoological and biomedical research.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Y.S.and G.Q.B.wrote the draft.All authors revised,read,and approved the final version of the manuscript.
Yan Shen1,Lu-Feng Ding1,Chao-Yu Yang1,2,Fang Xu1,Pak-Ming Lau1,2,3,Guo-Qiang Bi1,2,3,*
1Interdisciplinary Center for Brain Information,Brain Cognition and Brain Disease Institute,Shenzhen Institute of Advanced Technology,Chinese Academy of Sciences;Shenzhen-HongKong Institute of Brain Science-Shenzhen Fundamental Research Institutions,Shenzhen,Guangdong518000,China
2Division of Life Sciences and Medicine,University of Science and Technology of China,Hefei,Anhui230001,China
3Institute of Artificial Intelligence,Hefei Comprehensive National Science Center,Hefei,Anhui230001,China
*Corresponding author,E-mail:gqbi@ustc.edu.cn
- Zoological Research的其它文章
- Zoological Research call for papers of Cavefish Special Issue
- Fuel source shift or cost reduction:Context-dependent adaptation strategies in closely related Neodon fuscus and Lasiopodomys brandtii against hypoxia
- Ecological study of cave nectar bats reveals low risk of direct transmission of bat viruses to humans
- Population and conservation status of a transboundary group of black snub-nosed monkeys (Rhinopithecus strykeri) between China and Myanmar
- Nucleus accumbens-linked executive control networks mediating reversal learning in tree shrew brain
- Europe vs.China:Pholcus (Araneae,Pholcidae) from Yanshan-Taihang Mountains confirms uneven distribution of spiders in Eurasia

