Dynamic transcriptional atlas of male germ cells during porcine puberty
DEAR EDITOR,
Spermatogenesis is the process by which male gametes are formed from spermatogonial stem cells (SSCs) and it is essential for the reliable transmission of genetic information between generations.After birth,prospermatogonia (ProSG)give rise to SSCs.While mouse spermatogenesis is relatively well studied,we are only just beginning to unravel this process in larger animals.Here,we analyzed key developmental transitions and differentiation trajectories by profiling neonatal,juvenile,and adult testes through single-cell sequencing(scRNA-seq).We found that SSCs were established at 30 days old,and that CDH1 was a novel cell surface marker for porcine ProSG and undifferentiated spermatogonia.In addition,integrated analysis revealed similarities between pig and human spermatogenesis.This study is the first to analyze the transcriptome of male germ cells during porcine testicular development through scRNA-seq,and further document the development and classification of porcine germ cells.This study advances our understanding of the timing of porcine testicular development and provides a foundational data resource with multiple modalities of analyses.
In mice,primary transitional prospermatogonia (T1-ProSG)are mitotically quiescent cells and begin to convert into secondary transitional (T2)-ProSG at postnatal day 1,which then resume proliferation and give rise to SSCs.T2-ProSG generate the “first wave of spermatogenesis” (Tan et al.,2020).Subsequently,self-renewing SSCs commence differentiation and produce successive and continuous waves of gametogenesis from 2–3 weeks after birth.
However,porcine ProSG do not generate the equivalent“first wave” of spermatogenesis,and proliferate continuously after birth,albeit at a different growth rate.Porcine ProSG migrate to the basement membrane of the seminiferous tubules and differentiate into SSCs during the first three months of birth (Tan et al.,2020).Furthermore,SSC markers are not the same between pigs and rodent models (Park et al.,2019;Zhang et al.,2020).Therefore,the mechanisms regulating the differentiation of porcine ProSG and SSC are not fully understood,and a dynamic transcriptional germ cell atlas of testicular development is not yet available.
In this study,we profiled 9 307 single-cell transcriptomes from the whole-testes of one infant (7 days old (D7)) and three juvenile (30,60,and 90 days old (D30,D60,and D90,respectively)) Guanzhong black pigs for comparison with our recently published adult (150 days old (D150)) data (Zhang et al.,2021).Histological examination was conducted to explore the changes in morphology and complete timeline of porcine germ cell development,which revealed a clearly defined lamina and lumen across the tubules of the D90 and D150 samples (Figure 1A).ProSG and spermatogonia were detected before D90,and spermatocytes and a lower proportion of spermatids began to appear at D90.
To minimize batch effects,the mutual nearest neighbor(MNN) approach was used to integrate all scRNA-seq libraries.Using the Leiden algorithm and t-distributed stochastic neighbor embedding (t-SNE),41 cell clusters were identified.The clusters were then differentiated by known markers (e.g.,DDX4,UCHL1,SYCP3,andTNP2)(Supplementary Figure S1A–E) into three major germ cell types,i.e.,ProSG &spermatogonia,spermatocytes,and spermatids (Figure 1B).The marker genes used to classify the germ cell subgroups were as follows:ProSG &spermatogonia(UCHL1andKIT),spermatocytes (DMC1andPIWIL1),and spermatids (TNP1andPRM1) (Supplementary Figure S1F).
To investigate the classification and transcriptomic features of porcine ProSG and spermatogonia,re-clustering analysis was conducted on the ProSG &spermatogonia cluster(Supplementary Figure S2A),with 18 cell clusters identified(Supplementary Figure S2B).Clustering analysis revealed nine cell types based on well-known markers of ProSG and spermatogonia (Figure 1C,D).Two types of ProSG were identified (T1-ProSG and T2-ProSG).PIWIL4andSOX17were highly expressed in T1-ProSG,whileETV4andPALLDwere highly expressed in T2-ProSG (Supplementary Figure S2C).These results are important for the future isolation,culture,and characterization of ProSG.
To explore changes in the ratios and differences in the proliferative capacities of T1-ProSG and T2-ProSG,immunofluorescence analysis was conducted using the ProSG markerDolichos biflorusagglutinin (DBA) and cell proliferation marker proliferating cell nuclear antigen (PCNA)(Supplementary Figure S2D).Localization of DBA+cells was used to distinguish T1-ProSG (seminiferous tubule center) and T2-ProSG (seminiferous tubule periphery).Cell ratios of T1-ProSG and T2-ProSG were counted during testicular development (Figure 1E),which indicated that ProSG migration was asynchronous and the ratio of T2-ProSG gradually increased from birth to puberty.The proliferation of T1-ProSG,albeit at low rate,is consistent with previous histological examination of the relative number of germ cells(Frana et al.,2000).However,our findings are in contrast with previous rodent studies showing that ProSG are quiescent or degenerate massively after birth (Law &Oatley,2020).The proliferative capacity of T2-ProSG was greater than that of T1-ProSG (Figure 1F).Between the two ProSG populations,52 genes were up-regulated in T1-ProSG and 141 genes were up-regulated in T2-ProSG (Supplementary Figure S2E).Gene Ontology (GO) analysis indicated that cellcycle activation was enriched in T2-ProSG (Supplementary Figure S2F).Genes up-regulated in T1-ProSG were involved in the cell migration pathway (CAP1,CRK,JAK1,MESP2,CTTN,EZR,andIQGAP1),suggesting that these cells exhibit a strong migration ability.Moreover,genes highly expressed in T2-ProSG were involved in the cell proliferation pathway(PCNA,BUB1,CDK9,FGF8,andNEK2) (Supplementary Figure S2G).Taken together,T1-ProSG and T2-ProSG were primarily found at D7 and D30,which advances our understanding of porcine male germ cell development at this stage,with a large reduction at D60 and rare detection at D90(Supplementary Figure S2H).The ProSG began to transform into undifferentiated spermatogonia at D30,whereas differentiated spermatogonia began to appear at D60.Thus,it would be interesting to define T1-and T2-ProSG from a functional perspective in future work.
The cell-cycle score was calculated according to previously reported S and G2/M phase-specific genes (Nestorowa et al.,2016) (Supplementary Figure S3B).According to marker gene expression and cell-cycle analysis,16 clusters were grouped into seven spermatogonia clusters,i.e.,Undiff1,Undiff2,Undiff3,E-diff1,E-diff2,Mid-diff,and Late-diff (Figure 1C).SSC-associated marker genes,such asID4,PAX7,FGFR3,ZBTB16,andUCHL1,were highly expressed in the Undiff1 subgroup.Interestingly,the expression levels ofPAX7andID4,which are SSC function markers,were reduced when Undiff1 transitioned to Undiff2 (Figure 1D).Furthermore,most germ cells in the Undiff1 subgroup were at the G1/G0 phase of the cell cycle (Supplementary Figure S3B),indicating quiescence.ZBTB16andUCHL1were highly expressed in the Undiff3 subgroup,whileID4,PAX7,FGFR3,andETV5showed lower expression levels (Figure 1D).In rodents,SSCs and progenitor spermatogonia are comprised of As,Apr,and chains of Aal spermatogonia.SSCs are a subset of the As population and strongly expressID4(Fayomi &Orwig,2018).Thus,we propose that Undiff1,Undiff2,and Undiff3 resemble As,Apr,and Aal spermatogonia,respectively.To the best of our knowledge,this is the first detailed classification of spermatogonia in large animals,thus providing a foundation for future characterization of spermatogonia.
Cells in the E-diff1 subgroup partially corresponded to Aal and A1–A4 spermatogonia,expressing differentiation markers(KITandDMRT1) and proliferation marker (PCNA),with lower expression levels of SSC markers (ZBTB16andETV5)(Figure 1D).E-diff2 corresponded to intermediate spermatogonia,with high expression ofKIT,CTCFL,andDMRT1.Mid-diff corresponded to B spermatogonia,with high expression ofSTRA8,a marker of differentiating spermatogonia.Late-diff corresponded to pre-leptotene spermatocytes,with high expression ofSPO11,SYCP3,andREC8.
The porcine spermatocytes were divided into Lep,Zyg,Pachy,and Dip &Sec subsets (Figure 1G;Supplementary Figure S4).Meiotic recombination-associated genesREC8andSPO11were highly expressed in Lep,whereas meiosisspecific cyclin geneCCNA1was highly expressed in Dip &Sec (Figure 1H).Spermatids were classified into nine subgroups,i.e.,Early-RS1,Early-RS2,Early-RS3,Mid-RS,Late-RS,Early-ES,Mid-ES,Late-ES,and Sperm (Figure 1I;Supplementary Figure S4).CD63andNME5were highly expressed in Early-RS1-3,Mid-RS,and Late-RS.SPAG6 plays a critical role in acrosome formation and motility,whereas transition nuclear protein 1 and 2 (TNP1/2) are required for packaging and condensation of sperm chromatin.Here,SPAG6andTNP1/2were up-regulated when Late-RS transitioned to Early-ES and were further enhanced during differentiation.Following that,PRM1andPRM2showed high expression in the Sperm subgroup (Figure 1J).To elucidate the complete developmental trajectory of porcine spermatogenesis,we re-clustered the abovementioned annotated cell clusters (Figure 1C,G,I).Results showed a contiguous trajectory of porcine germ cell development,which allowed us to distinguish detailed changes that occurred during spermatogenesis (Figure 1K).Furthermore,pseudotime analysis was conducted to obtain an unbiased developmental trajectory of porcine spermatogenesis (Figure 1L).We thus proposed a model of porcine germ cell development (Figure 1O).Based on scRNA-seq analysis,partition-based graph abstraction (PAGA),and evidence from stage-specific genes,we reconstructed the developmental dynamics of porcine spermatogenesis and provided a molecular atlas of testicular development in pigs.
Interestingly,scRNA-seq analysis revealed that expression ofCDH1was up-regulated in ProSG (Figure 1D).Double immunofluorescence staining for CDH1/DBA and CDH1/UCHL1 revealed that most CDH1-positive cells overlapped with DBA-and UCHL1-positive cells(Supplementary Figure S5),indicating that CDH1-positive cells represent a subset of ProSG and undifferentiated spermatogonia in pigs.Therefore,CDH1 could be beneficial for sorting ProSG in neonate piglets,and potentially in other farm animals.Further studies are needed to uncover the role ofCDH1during germ cell proliferation and differentiation.
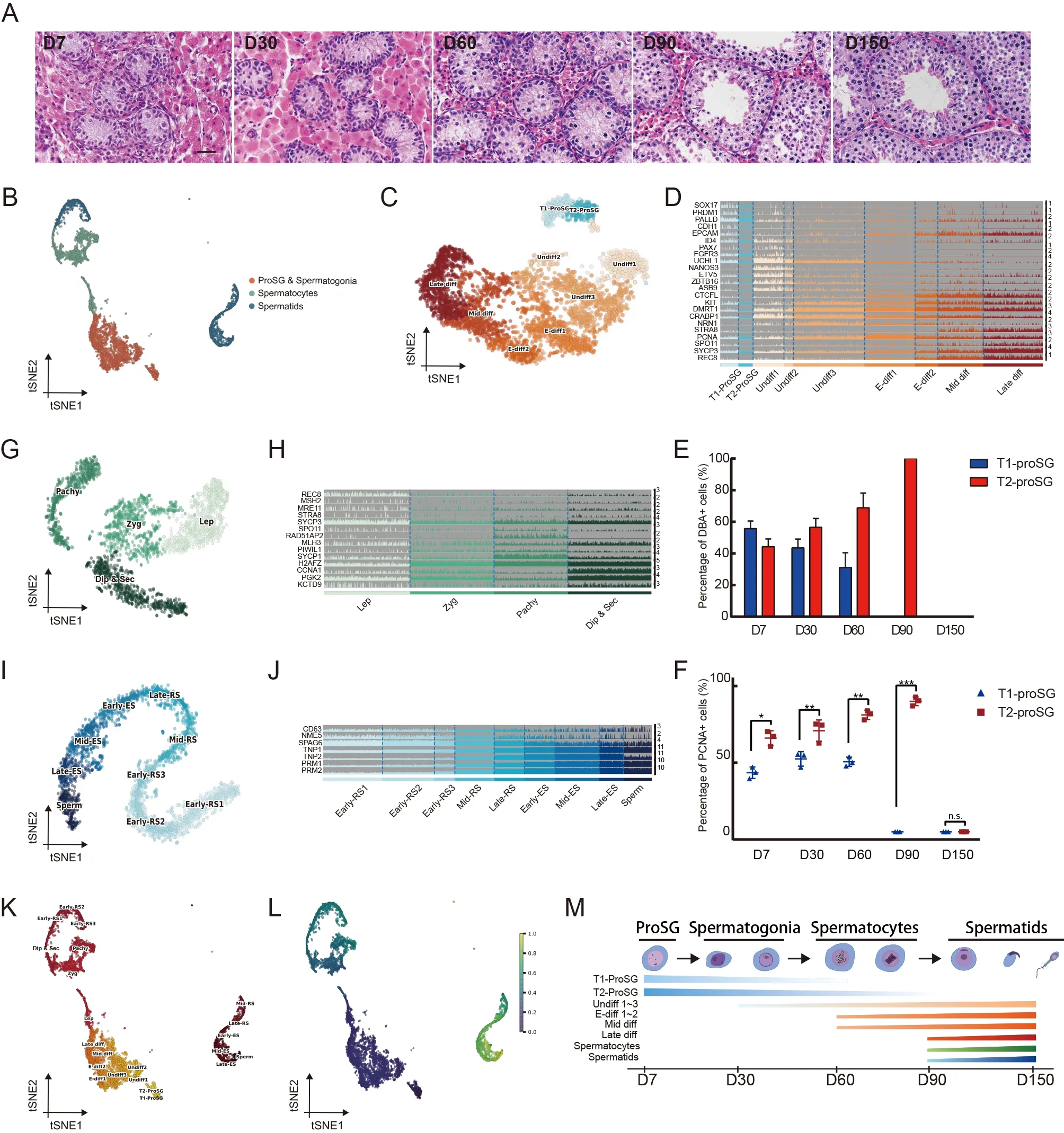
Figure 1 Single-cell RNA sequencing reveals atlas of germ cell development in porcine testes
During male meiosis,sex chromosomes undergo a tightly coordinated process known as meiotic sex chromosome inactivation (MSCI) (Turner,2007).The inactivation and reactivation of sex chromosomes were evaluated by plotting the mean number of genes expressed from the sex chromosomes compared to all autosomes (Supplementary Figure S6A).Genes of the X chromosome declined sharply at the Zyg stage(Supplementary Figure S6B).SCML2,which is critical for heterochromatin organization in MSCI,was highly expressed in the Lep stage (Supplementary Figure S6C).In addition,gene repression during MSCI was mediated by H3K9me3,an epigenetic marker catalyzed bySETDB1.Analysis revealed thatSETDB1was up-regulated in the spermatocytes(Supplementary Figure S6C),as verified by immunohistochemical assay (Supplementary Figure S6D).
Mammalian spermatogenesis is conserved among species and plays a crucial role in the transmission of genetic heritage.Pigs are important farm animals and one of the most important large animal models in biomedical research (Perleberg et al.,2018).In this study,comprehensive analysis revealed significant similarities in the spermatogenesis process between pigs and humans (Supplementary Figure S7).Our results and those of others (Fayomi &Orwig,2018) suggest that many traits are conserved across species.
In summary,we revealed key developmental transitions in differentiation trajectories by profiling neonatal,juvenile,and adult testes using 10× Genomics scRNA-seq.We identified two types of prospermatogonia,seven types of spermatogonia,four types of spermatocytes,and nine types of spermatids.We also identified CDH1 as a novel cell surface marker for ProSG and undifferentiated spermatogonia in pigs.Furthermore,our integrated analyses highlighted the similarities in porcine and human spermatogenesis.This study advances our understanding of testicular development from birth to puberty and provides a foundational resource with multiple modalities of analysis.
DATA AVAILABILITY
Datasets generated in this study are available in the GSA at the National Genomics Data Center (CRA010185),NCBI Gene Expression Omnibus (GSE186479) and Science Data Bank (https://www.scidb.cn/en/s/Zb6bMz).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
T.Z.and W.X.Z conceived and supervised the study.L.K.Z.analyzed the data.M.G.,H.D.M.,L.W.,Y.Z.,X.D.W.,T.J.L.,and H.Z.L.performed the experiments.T.Z.provided funding support.L.K.Z.,H.D.M,T.Z.,and W.X.Z.wrote the manuscript.All authors read and approved the final version of the manuscript.
Ling-Kai Zhang1,2,#,Hai-Dong Ma1,#,Ming Guo2,Ling Wang1,Yi Zheng2,Xiao-Dong Wu2,Tian-Jiao Li2,Hong-Zhao Lu1,Wen-Xian Zeng2,*,Tao Zhang1,*
1School of Biological Science and Engineering,Shaanxi University of Technology,Hanzhong,Shaanxi723001,China
2College of Animal Science and Technology,Northwest A&F University,Yangling,Shaanxi 712100,China
#Authors contributed equally to this work
*Corresponding authors,E-mail:zengwenxian2015@126.com;zhangtao780823@snut.edu.cn
- Zoological Research的其它文章
- Zoological Research call for papers of Cavefish Special Issue
- Fuel source shift or cost reduction:Context-dependent adaptation strategies in closely related Neodon fuscus and Lasiopodomys brandtii against hypoxia
- Ecological study of cave nectar bats reveals low risk of direct transmission of bat viruses to humans
- Population and conservation status of a transboundary group of black snub-nosed monkeys (Rhinopithecus strykeri) between China and Myanmar
- Nucleus accumbens-linked executive control networks mediating reversal learning in tree shrew brain
- Europe vs.China:Pholcus (Araneae,Pholcidae) from Yanshan-Taihang Mountains confirms uneven distribution of spiders in Eurasia

