Single-cell transcriptomic landscape of the sheep rumen provides insights into physiological programming development and adaptation of digestive strategies
Yuan Yuan ,Da-Ming Sun ,Tao Qin ,Sheng-Yong Mao ,Wei-Yun Zhu ,Yu-Yang Yin ,Jie Huang,Rasmus Heller,Zhi-Peng Li,Jun-Hua Liu,*,Qiang Qiu
1 School of Ecology and Environment, Northwestern Polytechnical University, Xi’an, Shaanxi 710072, China
2 Ruminant Nutrition and Feed Engineering Technology Research Center, College of Animal Science and Technology, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China
3 Laboratory of Gastrointestinal Microbiology, Jiangsu Key Laboratory of Gastrointestinal Nutrition and Animal Health, National Center for International Research on Animal Gut Nutrition, College of Animal Science and Technology, Nanjing Agricultural University, Nanjing,Jiangsu 210095, China
4 Huzhou Academy of Agricultural Sciences, Huzhou, Zhejiang 313000, China
5 Section for Computational and RNA Biology, Department of Biology, University of Copenhagen, Copenhagen DK-2200, Denmark
6 College of Animal Science and Technology, Jilin Agricultural University, Changchun, Jilin 130118, China
ABSTRACT As an important evolutionary innovation and unique organ,the rumen has played a crucial role in ruminant adaptation to complex ecological environments.However,the cellular basis of its complex morphology and function remains largely unknown.In this study,we identified eight major cell types from seven representative prenatal and postnatal rumen samples using~56 600 single-cell transcriptomes.We captured the dynamic changes and high heterogeneity in cellular and molecular profiles before,during,and after the appearance of keratinized stratified squamous epithelium with neatly arranged papillae and functional maturity.Basal cells,keratinocytes,differentiating keratinocytes,terminally differentiated keratinocytes,and special spinous cells provided the cellular basis for rumen epithelium formation.Notably,we obtained clear evidence of two keratinization processes involved in early papillogenesis and papillae keratinization and identified TBX3 as a potential marker gene.Importantly,enriched stratum spinosum cells played crucial roles in volatile fatty acid (VFA) metabolism and immune response.Our results provide a comprehensive transcriptional landscape of rumen development at single-cell resolution,as well as valuable insight into the interactions between dietary metabolism and the rumen.
Keywords:Rumen; scRNA-seq; Ruminal epithelium;Keratinization
INTRODUCTION
Ruminants are among the most successful mammalian lineages in animal evolutionary history (Chen et al.,2019).They possess remarkable and distinct multi-chambered stomachs,comprised of the rumen,reticulum,omasum,and abomasum,which collectively form a highly sophisticated natural bioreactor (Lei et al.,2018).This arrangement allows ruminants to utilize plant fiber more efficiently than other mammals,which may have led to their increasing success as extensive swathes of forest were gradually replaced by grassland around 40–50 million years ago (Chen et al.,2019).Consumed plant material is degraded by the dense and diverse microbiota in the rumen into precursors of essential metabolites and metabolic cofactors (particularly volatile fatty acids (VFAs),microbial proteins,and vitamins) used for the production of high-value meat and milk (Xu et al.,2021).Thus,the rumen and associated microbiota play key roles not only in ruminant growth,survival,and reproduction,but also in the provision of highly nutritious food for many human societies(Mizrahi &Jami,2018).In this symbiotic system,rumen tissue has evolved a series of distinct anatomical features and innovative functions that enable adaptation to the vast,complex rumen microbial ecosystem (Fonty et al.,1991;Russell &Rychlik,2001).Therefore,elucidating the cellular biological mechanisms underlying rumen formation and development is crucial for understanding the origin and evolution of the organ.
Unlike the stomachs of humans and monogastric animals(with simple,glandular,and prismatic epithelium),rumen tissue has a complex structure consisting of stratified squamous epithelium,lamina propria,mucosa,tunica muscularis,and plasma membrane (Groenewald,1993;Steven et al.,1970).In contrast to the smooth,non-keratinized stratified squamous epithelium of the esophagus (Oshima et al.,2011),the stratified squamous epithelium of the rumen,including the stratum corneum,granulosum,spinosum,and basal layer,is keratinized like skin epidermis (Graham &Simmons,2005).This notable structural feature of the rumen not only plays a protective role in defense against microbial invasion but also facilitates renewal of rumen tissue (Graham&Simmons,2005).This suggests that immunoregulation in the rumen may be involved in maintaining homeostasis,although the idea of the rumen as an immune-related organ remains controversial.Previous transcriptomic analyses have suggested that theKRT4gene and keratinocyte differentiation pathway play key roles in stratum corneum formation (Bragulla&Homberger,2009;Pan et al.,2020).The stratified squamous epithelium is the main site of VFA metabolism and ketogenesis in adult ruminants,thereby affecting the availability of energy and nutrients for the host (Rémond et al.,1995).Immunohistochemical and morphometric analyses have indicated that Na+-K+-ATPase α-subunit expression and mitochondrial density and distribution in the stratum spinosum and basal layer of the rumen epithelium facilitate VFA metabolism (Giesecke et al.,1979;Graham &Simmons,2005).In addition,the rumen contains diverse and interdependent populations of bacteria,archaea,protozoa,fungi,and viruses (Russell &Rychlik,2001).However,the cellular basis of its complex morphological structure and innovative functions remains largely unknown.
Studying the development of an organ is an effective way to understand its cellular biological foundation (Finnegan et al.,2019;Haber et al.,2017).Morphometric analysis has shown that embryonic sheep rumen tissue forms three differentiated layers at~42 days of gestation,with the papillary structure and keratinized stratified squamous epithelium forming at~57 days of gestation (Franco et al.,2011).Postnatal rumen development differs from that of the stomach in monogastric animals.In neonatal ruminants,the fluid (milk or milk replacer)obtained from suckling primarily flows into the abomasum via the esophageal groove in the rumen,resulting in a lack of rumen microbial fermentation substrates to stimulate rumen development (Beharka et al.,1998;Wise &Anderson,1939).After the introduction of solid food,rumen papillae continue to grow and the VFA metabolism machinery gradually matures(Baldwin &Connor,2017;Pazoki et al.,2017;Rémond et al.,1995).These findings suggest that dramatic structural and functional changes occur during embryonic and postnatal rumen development,implying that cell type and composition have significantly changed.However,the critical cellular processes that occur during these developmental periods remain unclear.
Here,we performed single-cell RNA sequencing (scRNAseq) to explore the cellular heterogeneity and function of specific cell types in rumen development.We profiled~56 600 single-cell transcriptomes from seven key points in sheep rumen development (spanning prenatal to postnatal stages) to address the following questions.First,which cells form the complex morphological structure of the rumen? Second,what processes are involved in the formation of each cell type?Third,which cells play fundamental roles in specific functions and features (keratinization,papilla formation,VFA metabolism,and immunological functions) of the rumen? Our findings should help elucidate the composition,development,and functional maintenance of the rumen at the cellular level and provide valuable insight into the interactions between dietary metabolism and rumen function.
MATERIALS AND METHODS
Sheep rumen dissection
The experimental design and procedures were approved by the Animal Care and Use Committee of Nanjing Agricultural University.Domesticated Hu sheep (a native breed in China with high reproductive efficiency and the ability to tolerate large amounts of roughage) were selected.Prenatal rumen samples were collected from healthy pregnant Hu sheep (at 30,60,90,110,and 130 days of gestation) via caesarean section,and postnatal rumen samples were obtained from newborn and 45-day-old lambs.Fetuses and lambs were stunned by electric shock and killed by exsanguination.The whole rumen was immediately excised and placed in ice-cold phosphate-buffered saline (PBS) (Sigma-Aldrich,USA),after which fatty tissue and blood vessels on the outer wall were carefully removed.
Primary cell isolation
Each rumen sample was incised along the dorsal curvature and washed in cold Hanks’ Balanced Salt Solution (HBSS,Gibco,USA) several times to remove mucus and blood.After collecting samples for morphological observation,the rumen tissue was cut into small pieces (5 mm),washed with HBSS until clear,and repeatedly digested with 0.25% trypsin (Gibco,USA) for 25 min at 37 °C.The digestion solution was changed every 5 min,collected in a centrifuge tube containing fetal calf serum,and filtered through a cell strainer (40 μm,Gibco,USA).After solution centrifugation (3 min at 1 500 r/min,4 °C),the sediment was resuspended in PBS to remove residual trypsin.The fetal rumen cells were centrifuged (3 min at 1 500 r/min,4 °C) again and resuspended in PBS to determine density and viability using a Bio-Rad TC20 Automated Cell Counter (Bio-Rad,USA).Cell viability was confirmed by trypan blue staining,and cell density was adjusted to 1 000 000 cells/mL.Finally,live cells were selected for scRNA-seq using the 10x Genomics single-cell platform.
ScRNA-seq data processing
Single-cell raw data were processed using 10x Cell Ranger software (v3.0),including “cellranger mkfastq” to demultiplex raw data into Fastq files and “cellranger count” for alignment,filtering,barcode counting,and unique molecular identifier(UMI) counting using default settings.A sheep reference genome (Oar_rambouillet_v1.0) was downloaded from ENSEMBL and built with “cellranger mkgtf”.Finally,we obtained seven UMI count tables (absolute numbers of observed transcripts) for further analysis,i.e.,one for each developmental stage.
Analysis and visualization of processed sequencing data
For downstream analyses,we retained cells expressing at least 200 genes but less than 25 mitochondrial genes.To obtain a single-cell expression matrix for the retained cells,we applied the “FindIntegrationAnchors” and “IntegrateData”functions in Seurat (v3.1.4) to merge the data and used the“ScaleData” function to regress out mitochondrial gene expression.We then used the “RunPCA” function in Seurat for principal component analysis (PCA) of the corrected cell expression matrix.The number of significant principal components (PCs) was determined using the permutationPA function in the jackstraw R package (https://cran.rproject.org/web/packages/jackstraw).Analysis identified 45 significant PCs,the scores of which were used for further analysis.We then applied the “FindClusters” function in Seurat for cell clustering analysis.We set the parameter resolution to 1.2 and identified 31 clusters.Uniform Manifold Approximation and Projection (UMAP) and t-distributed Stochastic Neighbor Embedding (t-SNE) plots of the second-level clustering results were obtained with Seurat,as above.
Identification of cell types and differentially expressed genes (DEGs)
Differences in gene expression among cell clusters were analysed using the “FindMarkers” function in Seurat.The marker genes visualized in Figure 1C were identified using the FindAllMarkers Seurat function with default settings.The marker genes were analyzed using David (https://david.ncifcrf.gov) for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses.
Single-cell pseudo-time trajectory analysis
Monocle (v2.14.0) was used for ordering single cells in pseudo-time (http://cole-trapnell-lab.github.io/monoclerelease/ docs/#constructing-single-cell-trajectories).We constructed single-cell differentiation trajectories using Monocle-identified variable genes as ordering genes.Root state was determined based on Seurat cell identity information.Gene clusters were further divided into four clusters according to theirk-means.To investigate gene function for each gene cluster,we performed GO and KEGG enrichment analyses using David (https://david.ncifcrf.gov).
Statistical analysis
Statistical analyses were performed using the R package.Pvalues were calculated using two-tailed Student’st-tests,with values below 0.05 considered statistically significant.
RESULTS AND DISCUSSION
Identification of diverse cell types in rumen
To investigate the key features of rumen development,we collected seven representative Hu sheep rumen samples from the prenatal stage to functional maturity:i.e.,at 30,60,90,110,and 130 days of gestation,newborn (at 144 days of gestation),and 45 days old (Figure 1A).We profiled~56 600 single-cell transcriptomes from these samples and partitioned the cells into 31 clusters (Figure 1B),each containing 194 to 4 291 cells.Based on the distinct expression patterns and levels of known cell-type marker genes (Figure 1C;Supplementary Table S1),we classified these clusters into eight cell types,including epithelial,fibroblast,smooth muscle,endothelial,proliferating,lymphocyte,macrophage,and T cells (Figure 1D).
Epithelial cells,the most variable cell population during rumen development,including basal cells,keratinocytes,differentiating keratinocytes (DKs),terminally differentiated keratinocytes (TDKs),and special spinous cells (SSCs),rapidly increased in proportion at 60 days of gestation(Figure 1E).Clusters 0,7,and 9 were classified as basal cells with high expression of marker genes such asKRT15andCOL17A1(Joost et al.,2016) (Figure 1C).Cluster 20 cells were annotated as keratinocytes with high expression ofJUN,FOS,andZFP36,which are predominantly expressed in keratinocytes and participate in differentiation (Mehic et al.,2005;Prenzler et al.,2016) (Figure 1C).Clusters 2,4,10,12,23,24,and 25 were classified as DKs with high expression ofFABP5,KRTDAP,SPINK5,andCSTA(Figure 1C),which play roles in keratinocyte differentiation (Finnegan et al.,2019;Ogawa et al.,2011).Clusters 14,15,and 16 were classified as TDKs defined by high expression ofCNFNandKRT23(Figure 1C),which are associated with the cornified cell envelope of the stratified squamous epithelium (Wagner et al.,2019).Clusters 1,3,5,6,and 13 were identified as SSCs with high expression of lipid metabolism and ion transport-related genesCA1,ACAA2,andBDH1(Bond et al.,2019)(Figure 1C).
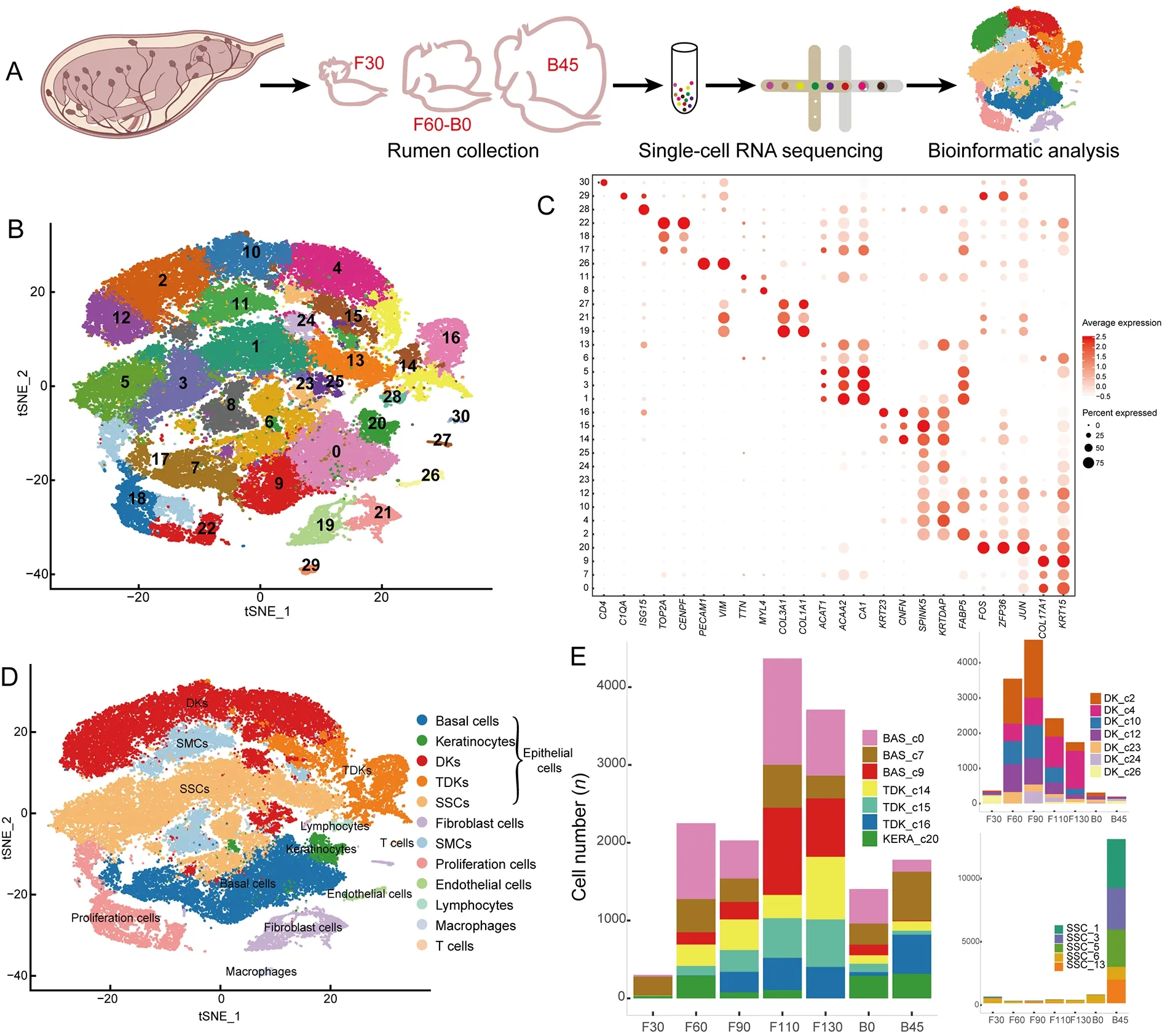
Figure 1 Single-cell transcriptome profiling of sheep rumen
In addition to epithelial cells,Clusters 19,21,and 27 were identified as fibroblasts with high expression of extracellular matrix (ECM) genes (COL3A1,COL1A1,POSTN,andDCN)(Figure 1C) (Han et al.,2020).Clusters 8 and 11 were annotated as smooth muscle cells (SMCs) with high expression ofACTC1,MYL4,andTTN(Han et al.,2020)(Figure 1C).Clusters 17,18,and 22 were identified as proliferating cells with high expression of cell cycle-related genes,such asCENPF,TOP2A,andHIST2H2AC(Han et al.,2020) (Figure 1C).Cluster 26 cells were annotated as endothelial cells with high expression ofVIMandPECAM1(Han et al.,2020) (Figure 1C).Cluster 28 cells were identified as lymphocytes with high expression ofISG15,MX1,andDDX58(Wani et al.,2019) (Figure 1C),Cluster 29 cells were identified as macrophages due to expression ofC1QA,C1QB,andC1QC(Cochain et al.,2018) (Figure 1C),and Cluster 30 cells were identified as T cells expressingCD4andCD69(Schoenberger,2012) (Figure 1C).
In summary,scRNA-seq successfully identified cell types of the rumen from the embryonic to functionally mature postnatal stages.This new comprehensive atlas of gene expression and marker genes in diverse cell clusters of the rumen provides a robust foundation for subsequent studies of rumen development (Table 1).
Diverse cell-type subpopulations in the rumen
Given its importance in diet metabolism and microbial defense,rumen development and functional maturation play crucial roles in ruminant physiology (Baldwin &Connor,2017).The rumen epithelium develops from two layers of cells at the embryonic state (i.e.,stratum basale and stratum granulosum)to a four-layer structure after birth (i.e.,stratum basale,stratum spinosum,stratum granulosum,and stratum corneum)(García et al.,2012).Based on the identification of diverse epithelial cell types,we further investigated the genetic characteristics of the four strata of the rumen epithelium.Specific clusters were defined by cell-type abbreviation and cluster number,e.g.,BAS-c0 refers to Cluster 0 (basal cells)and DK_c24 refers to Cluster 24 (differentiating keratinocytes).Basal cells are the main cells in the stratum basale (Jenkins&Tortora,2016).We identified three basal cell subpopulations(BAS_c0,BAS_c7,and BAS_c9;Figure 2A),which showed high expression of insulin-like growth factor binding proteinrelated genes (IGFBP2,IGFBP5,andIGFBP6),with particularly strong expression in BAS_c0 and BAS_c9(Figure 2A;Supplementary Table S2).These genes participate in epithelial-mesenchymal transition (EMT) (Gao et al.,2016;Vijayan et al.,2013),implying that EMT is essential for rumen development at the embryo stage.Furthermore,BAS_c7 cells showed high expression ofHELLSandUHRF1(Figure 2A),which play important roles in epidermal homeostasis (Wang et al.,2020),and ofFABP5,which participates in keratinocyte differentiation (Ogawa et al.,2011).The proportions of the BAS_c0 and BAS_c9 cells were lower at postnatal day 45 (Figure 1E).We speculate that BAS_c0 and BAS_c9 cells are associated with overall rumen development during the embryonic period and BAS_c7 cells are primarily associated with development of the rumen epithelium.
Keratinocytes are the main cells involved in keratinization(Candi et al.,2005).In addition to keratinocytes (KERA_c20),we identified seven DK subpopulations (DK_c2,DK_c4,DK_c10,DK_c12,DK_c23,DK_c24,and DK_c25) and three TDK subpopulations (TDK_c14,TDK_c15,and TDK_c16) in the rumen (Figure 2B).The relative proportions of DKs and TDKs remained relatively stable throughout the embryonic period (Figure 1E).However,the relative proportions of DK_c2 and DK_c12 decreased,while the relative proportions of DK_c4 and TDK populations increased with development of the embryonic rumen.KEGG pathway enrichment analysis indicated that up-regulated DEGs in KERA_c20 cells were enriched in the tumor necrosis factor (TNF) signaling pathway(Supplementary Table S3),which is involved in cell survival,death,and differentiation (Webster &Vucic,2020),thus suggesting that it may be crucial for keratinocyte differentiation.Comparison of DEGs (Supplementary Table S4) in the DK subpopulations showed that DK_c2 and DK_c10 cells highly expressedFABP5andTFF3(Figure 2B).FABP5induces keratinocyte differentiation through regulation of fatty acid transport (Ogawa et al.,2011),whileTFF3is a modifying factor in pathways regulating cell proliferation and keratinocyte migration (Storesund et al.,2009).Therefore,these two cell types may be associated with keratinocyte proliferation and differentiation.The DK_c4 and DK_c24 cells highly expressedANXA1andCSTA(Figure 2B),which participate in the cornified envelope formation in keratinocytes (Ma &Ozers,1996;Steven &Steinert,1994),suggesting that these two subpopulations may be associated with keratinization.The three TDK subpopulations (TDK_c14,TDK_c15,and TDK_c16) showed different gene expression profiles(Figure 3A;Supplementary Table S5).TDK_c14 cells showed high expression of ribosomal protein genes.TDK_c15 cells showed high expression ofAHNAK,DSP,andJUP,which are associated with desmoplastic plaques (Masunaga et al.,1995;Yang et al.,2006),indicating a contribution of TDK_c15 to cell adhesion and maintenance of ruminal epithelium stability.TDK_c16 cells showed high expression of innate immunerelated genes (IRF7,ISG15,FADD,MAPK14,andMAPK13),suggesting that the stratum corneum participates in innate immunity in addition to physical defense.

Figure 2 Characteristics of cell composition and gene expression patterns in the rumen
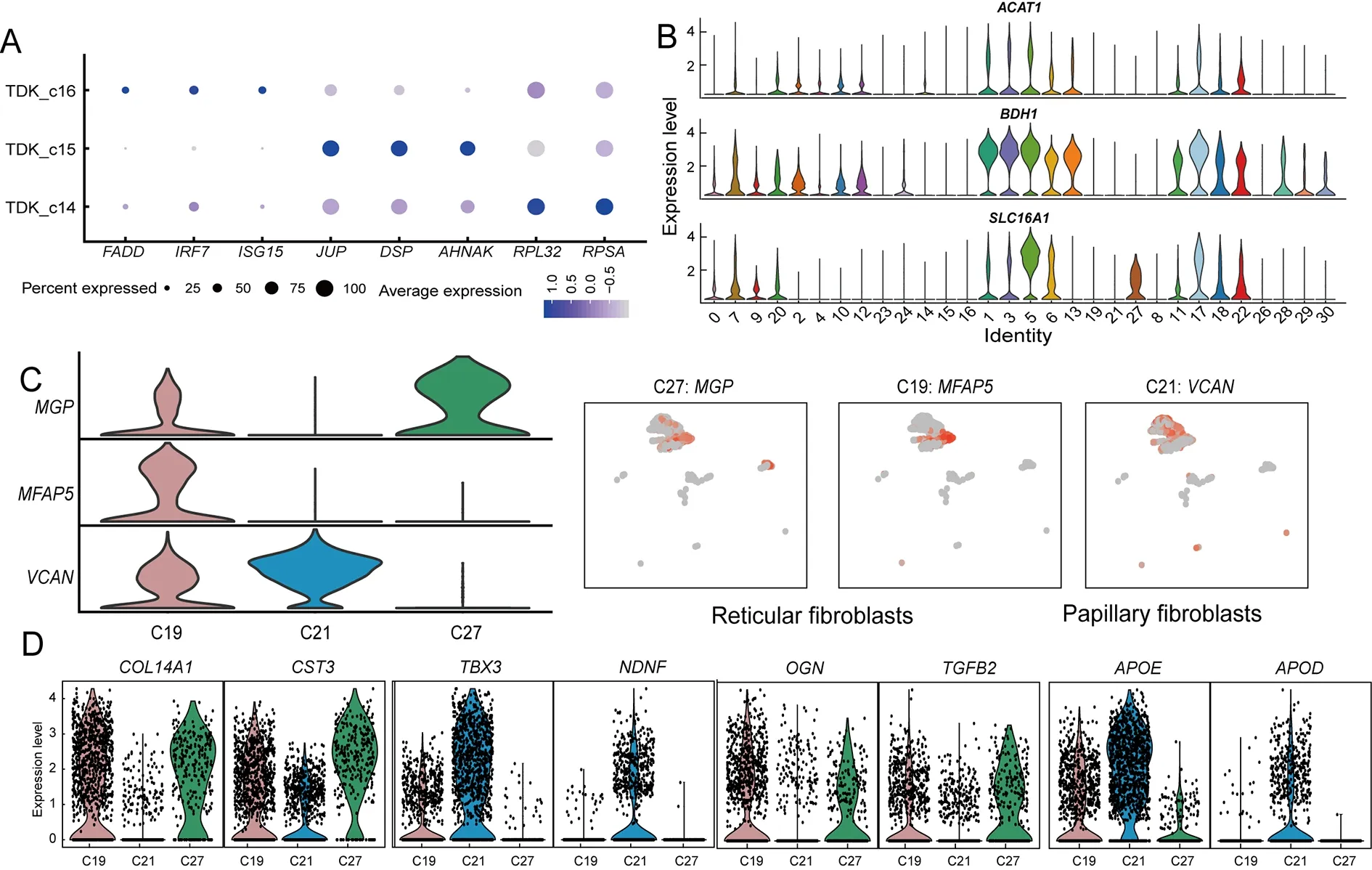
Figure 3 Expression of selected genes across rumen cell populations
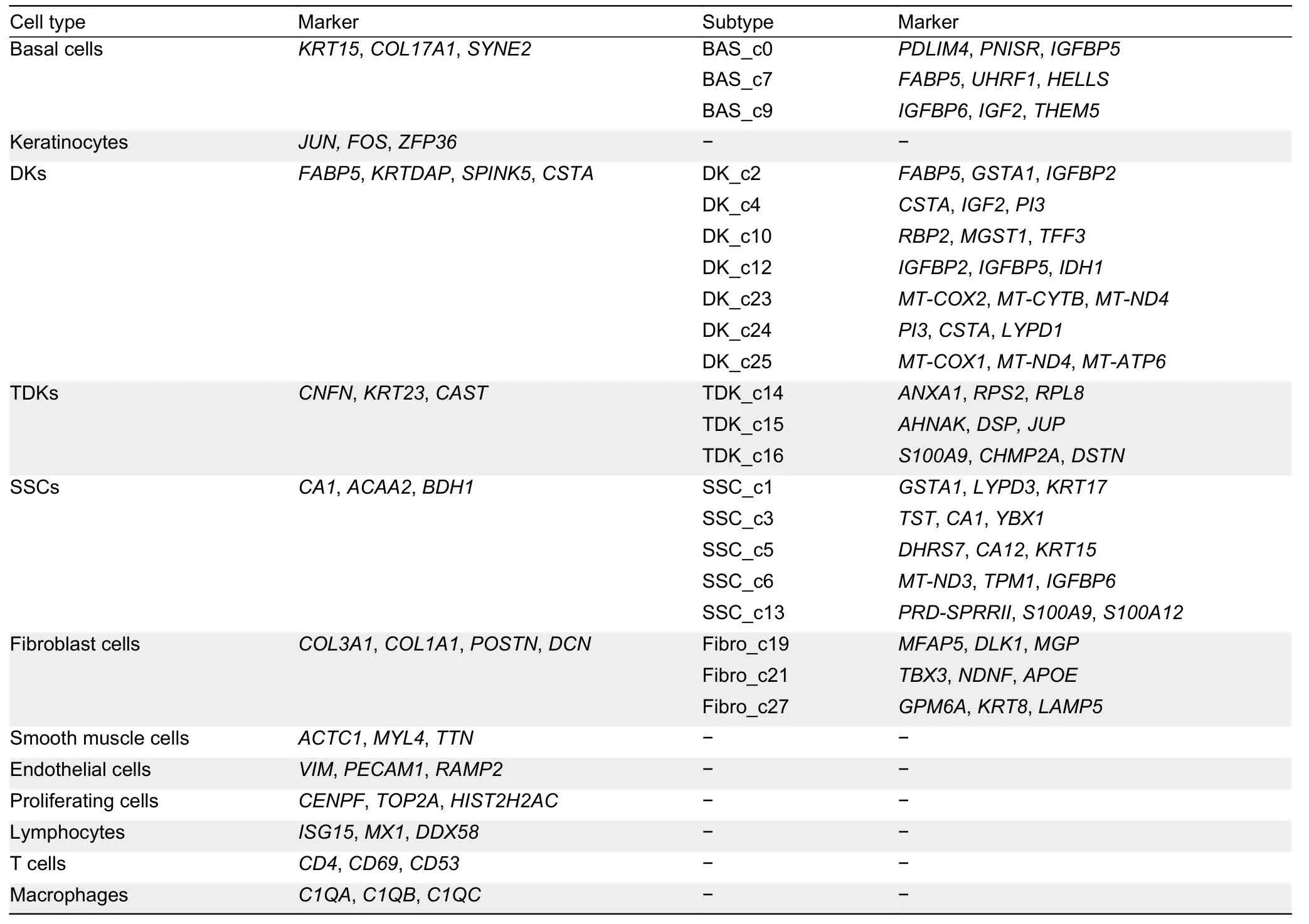
Table 1 Summary of marker genes of each cell type identified in rumen
The stratum spinosum is the main metabolic layer of the rumen and can be distinguished morphologically after birth(Baldwin &Connor,2017).Comparison of the proportions of epithelial cells in different periods showed that five clusters(proliferating cell cluster PC_c17 and SSC clusters SSC_c1,SSC_c3,SSC_c5,and SSC_c13) were significantly more abundant at postnatal day 45 (Figure 1E).These five subpopulations showed high expression of key ketogenesisrelated genes (ACAT1,SLC16A1,andBDH1) (Figure 3B;Supplementary Table S6).Ketogenesis is a hallmark of metabolic development of the rumen epithelium (Naeem et al.,2012).Therefore,we defined these five cell clusters as the major cell types of the rumen epithelium stratum spinosum.Among the SSC subpopulations,SSC_c5 cells exhibited high expression of metabolic genes and marker genes of basal cells (KRT15andIGFBP2) (Figure 2C),suggesting that they have a similar differentiation capacity to basal cells.SSC_c3 cells showed high expression of mitochondrial genes,suggesting a major role in energy metabolism.SSC_c1 and SSC_c13 cells showed high expression ofSPINK5andPRDSPRRII,indicating that these two subpopulations exhibited the highest degree of keratinization.Interestingly,PC_c17 cells expressed more ketogenesis-related genes than other proliferating cells (PC_c18 and PC_c22),suggesting that PC_c17 may represent the proliferative state of SSCs(Figures 2C,3B).Thus,we hypothesize that the SSC and PC_17 cell subpopulations may play key roles in the renewal and proliferation of the stratum spinosum,thereby maintaining the homeostasis of rumen metabolic function.
Fibroblasts,which are enriched in the rumen lamina propria,not only produce extracellular matrix (ECM) to provide structural support for the epithelium,but also participate in cell signaling,migration,proliferation,and differentiation (Kim et al.,2011).We identified three types of fibroblasts (Clusters 19,21,and 27) in the rumen (Figure 2D).Both C19 and C27 cells showed high expression ofCOL14A1(collagen-encoding gene) andMGPin comparison with C21 but could be distinguished by differential expression ofMFAP5andKRT8(Figure 3C).C21 cells strongly expressed lipid transportrelated genesAPOEandAPODand collagen-encoding geneCOL13A1(Figure 3D).Comparison of the DEGs in the three fibroblast types indicated that C21 consisted of papillary fibroblasts,while C19 and C27 consisted of reticular fibroblasts.We further found that the papillary fibroblasts(C21) strongly expressed neuro-development (NDNF)-related genes,suggesting that papillary fibroblasts also participate in rumen innervation (Figure 3D).
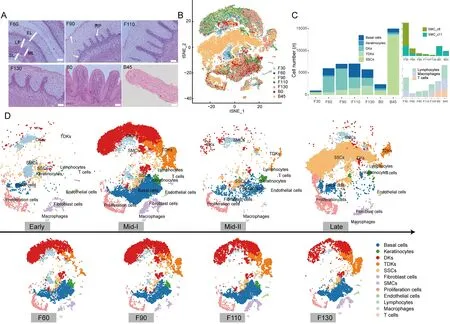
Figure 4 Characterization of rumen developmental patterns
In summary,combined with the characteristics of the rumen structure,we identified specific cell types associated with the morphological strata of the rumen and the heterogeneity of cells in these strata.Different cell subpopulations showed specific gene expression characteristics and different functions during rumen development.
Characterization of dynamic prenatal and postnatal rumen developmental patterns
Based on morphometric analysis (Figure 4A) and expression characteristics of samples obtained from seven periods of rumen development (Figure 4B;Supplementary Table S7),we distinguished four major stages of development,i.e.,early(0–30 days of gestation),middle-I (60,90,110,and 130 days of gestation),middle-II (newborn),and late (45 days old)(Figure 4D).
During the early stage of rumen development,the rumen becomes a small separate compartment of the primitive gastric tube.The ruminal wall consists of four layers:i.e.,internal epithelial layer,middle pluripotential blastemic tissue layer,tunica muscularis (with an inner circular layer and outer longitudinal layer),and serosa (García et al.,2012;Jenkins &Tortora,2016).Cell-type analysis showed that all cell types were present in the early stage,but SMCs were dominant(Figure 4C).We identified two SMC clusters (SMC_c8 and SMC_c11),which corresponded to the inner circular layer and outer longitudinal layer of the tunica muscularis (Martini,2006).In addition,scRNA-seq confirmed the presence of immune cells (lymphocytes,macrophages,and T cells) in the early-stage rumen (Figure 4C),which has been controversial due to their small numbers and difficult detection by conventional methods.
In the middle-I stage of rumen development,the ruminal wall consisted of four layers:i.e.,epithelium,lamina propria +submucosa,tunica muscularis,and serosa (Figure 4A).Morphological changed include ruminal papillogenesis and maturation of the epithelial layer (Figure 4A).With the formation of the rumen epithelium,several epithelial cells(basal cells,keratinocytes,DKs,TDKs) became the dominant type in the rumen,and the proportions of all epithelial cell types increased significantly at 60 days of gestation(Figure 4C).We also observed significant changes in the proportions of various epithelial cell types with increasing gestation time (Figure 1E,4C).The percentages of keratinocytes and TDKs decreased and increased with gestation time,respectively (Figure 4C),coinciding with gradual keratinization during rumen development (Figure 4A).Furthermore,at 90 days of gestation,we found that early epithelial stratification was accompanied by slight elongation of the stratum germinativum into the stratum granulosum,forming rudimentary ruminal papillae (Figure 4A).Accordingly,the proportion of DK_c24 cells was higher at 90 days than at 60 days of gestation,suggesting that these cells may be associated with early papillogenesis.Hence,the dynamic changes in different epithelial cell types provide the cellular basis for the specific morphology of the mature rumen(keratinization,papilla formation) at this developmental stage.
In the middle-II stage,the rumen papilla structure formed and the epithelium differentiated into four layers:i.e.,stratum corneum,stratum granulosum,stratum spinosum,and stratum basale (Figure 4A).We found that the proportions of basal cells,keratinocytes,TDKs,and SSCs in this stage were relatively balanced,while the proportions of DKs and fibroblasts decreased.In addition,the proportion of SSCs,major contributors to the metabolic capacity of the rumen,highly increased (Figure 4C),indicating that newborns formed a metabolically competent rumen before receiving external food stimulation,although SSCs were not the dominant cell type at this stage.
In the late stage of rumen development,SSCs became the dominant cell type,accounting for~85% of the rumen cell population,and proliferating cells increased significantly(Supplementary Table S8).These findings suggest that dramatic changes in rumen cell composition are adaptive responses to the external environment after solid diet introduction.Overall,these results reveal the dynamic changes in cell type and proportion that occur during rumen development in sheep,providing valuable clues for large-scale studies of rumen development in ruminants.
Characteristics of keratinization and papillogenesis during rumen development
Ruminal parakeratosis is a common ruminant disease caused by abnormal keratinization (Underwood et al.,2015),which impairs ruminant health and production by affecting defense and absorption of the rumen epithelium (Steele et al.,2009).We explored rumen keratinization in the current study.Based on pseudo-time analysis (Guo et al.,2021),we identified two distinct keratinization processes during normal rumen development (Figure 5A).One process (designated P1)involved progression through basal,KERA_c20,DK_c12,DK_c2,DK_c10,DK_c4,and TDK cellular states.The other process (designated P2) involved progression through basal,KERA_c20,DK_c12,DK_c2,DK_c10,DK_c24,DK_c4,and TDK cellular states (Figure 5B).These two keratinization processes exhibited similar dynamic gene expression profiles(Figure 5C,D),although passage through the DK_c24 state was only observed in P2.The DK_c24 cells showed high expression ofPI3(Supplementary Table S4),which is associated with elongation of the lateral cell membrane (Wang&Brieher,2020),and a rapid increase in abundance in the early papillogenesis phase at prenatal day 90.We hypothesize that DK_c24 cells are a transitional state for keratinocytes that appear during early papillogenesis of the rumen,and P2 is the process of papillae keratinization.
To further elucidate the P2 keratinization process,we visualized dynamic gene expression patterns along the pseudo-time trajectory and classified them into four groups(Figure 5C;Supplementary Table S9).Group 1 contained 317 genes highly expressed in early keratinization,includingSOX2,MEIS1,RIF1,APC,ID3,SMARCAD1,SMAD5,andFGFR2(Figure 5E),which are associated with regulation of stem cell pluripotency (Kanehisa et al.,2021).These genes were enriched in the GO terms “nucleoplasm” and “DNA binding”,suggesting that cell differentiation is a major step in early keratinization (Supplementary Table S10).The main keratin-encoding gene in this group wasKRT17(Figure 5E).Group 2 included 364 genes highly expressed in the midstage of keratinization,including genes associated with mitochondrial formation,maintenance,and function.These genes were enriched in the “Ribosome” pathway,indicating high rates of protein synthesis (Supplementary Table S10).The main keratin-encoding gene expressed in this group wasKRT15(Figure 5E).Group 3 contained 306 genes highly expressed in the mid-stage of keratinization,includingKRT4,ANXA7,VDAC1,andCNN3(Figure 5E),which are associated with epithelial cell differentiation (Kanehisa et al.,2021).These genes were enriched in the GO terms “microtubule”and “focal adhesion” (Supplementary Table S10),suggesting that cellular connections are enhanced during this stage of keratinization.Group 4 contained 1 242 genes highly expressed in late keratinization and enriched in the KEGG terms “adhesion junction” and “bacterial invasion of epithelial cells” (Supplementary Table S10).Genes of the S100A family(S100A2,S100A4,S100A5,S100A9,S100A10,S100A12,andS100A13),which are associated with the immune system(Xia et al.,2018),were also highly expressed in the late stage(Figure 5E),indicating that the rumen epithelium develops defensive capacities in this stage.

Figure 5 Molecular features of keratinization and papillogenesis during rumen development
The neatly arranged papillae are a striking morphological feature of the mature rumen,enlarging the surface area available for nutrient absorption and metabolism (Graham &Simmons,2005).Anormogenesis of rumen papillae can trigger subacute ruminal acidosis and other nutritional metabolic diseases that can severely impair animal production and health (Underwood et al.,2015).Papillary fibroblasts,which are close to the epithelial layer and associated with papilla formation,showed higherTBX3transcription factor expression than the reticular fibroblasts (Figure 5F).Previous studies have shown that abnormal expression ofTBX3in humans can cause papillary carcinoma of the skin,ulnarmammary syndrome,and arrhythmia (Frank et al.,2012;Khan et al.,2020).HighTBX3expression can also promote expression ofPI3(Peres et al.,2015),which was strongly expressed in the papillogenesis-related DK_c24 cells.We speculate that the increased expression ofTBX3in the papillary fibroblasts may promote an increase inPI3expression in the DK_c24 cells,leading to elongation of the lateral membrane of the epithelial cells and hence papillogenesis (Figure 5G).In addition,we found that the five cell clusters in the stratum spinosum highly expressedKRT36(Figure 5H),a specific marker of filiform papillae (Brychtova et al.,2020).This suggests that stratum spinosum cells are in the papillae and corroborates the importance of rumen papillae as sites of energy metabolism.
Functional rumen epithelium formation after birth
With the introduction of a solid diet after birth,the proportion of SSCs (1,3,5,and 13) and PC_c17 cells increased and became dominant in the rumen epithelium and formed the stratum spinosum by postnatal day 45.We identified 407 genes highly expressed in SSCs.KEGG enrichment analysis of up-regulated DEGs revealed that 73 genes were enriched in metabolic pathways,including carbon metabolism,fatty acid metabolism,propanoate metabolism,and butanoate metabolism (Supplementary Tables S11,S12).These findings suggest that the major cellular components required for a functionally mature rumen epithelium are established before postnatal day 45.
As ruminants are exposed to a highly complex environment from birth,the rumen must not only develop physical defenses,but also an immune response.However,there were no significant differences in the expression levels of immunerelated genes among the three types of immune cells in the rumen before and after birth (Figure 6A).Interestingly,we found that SSC_c13 was the most keratinized subpopulation of SSCs,and cells in this cluster showed high expression of immune-related genes (ISG15,IL36A,and S100A gene family) (Figure 6B).ISG15plays a key role in the innate immune system,inducing natural killer cell proliferation (Perng&Lenschow,2018).ISG15also induces interferon (IFN)cytokines as an anti-mycobacterial response and is highly expressed in lymphocytes (Gao et al.,2020).Thus,we hypothesize that SSC_c15 cells induce lymphocyte proliferation to generate an immune response.These findings suggest that maturation of the postnatal rumen stratum spinosum is not only important for ruminant energy metabolism,but also for immune capacity.
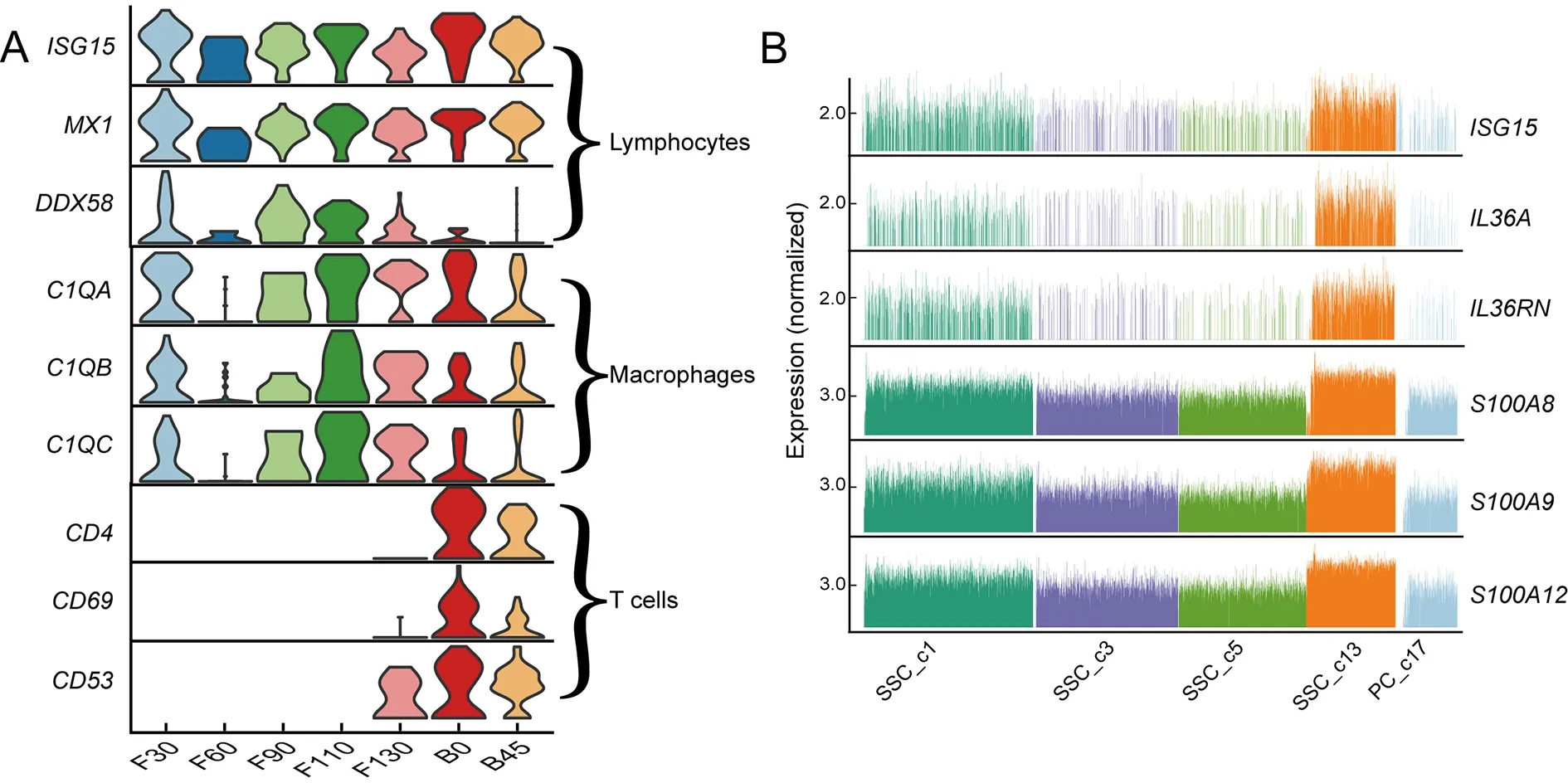
Figure 6 Molecular features of various cell types in rumen of lambs after birth
CONCLUSIONS
As an important evolutionary innovation and unique organ,the rumen has played an important role in the adaptation of ruminants to their complex ecological environments.This study provides a detailed account of gene expression during prenatal to postnatal rumen development based on scRNAseq analysis,including detailed characterization of the cell types involved,as well as identification of additional marker genes for specific cell types.We revealed the dynamic changes and high heterogeneity of the cellular and molecular profiles during four development stages of the rumen,providing clues for maternal regulation of fetal rumen development.However,this study is limited by the small sample size and number of captured cells.
The rumen has evolved a series of distinct structural hallmarks,such as neatly arranged papillae and keratinized stratified squamous epithelium,as well as innovative functional characteristics,such as VFA metabolism and immunology.Our identification of diverse basal cells,keratinocytes,DKs,TDKs,and SSCs provides important insight into rumen structure and function and a basis for isolating different cells in the rumen.The epithelial cells exhibit two processes during papillary keratinization accompanied by dynamic changes in identified gene expression.In addition,the cellular and gene expression profiles strongly suggested that the rumen epithelium is functionally mature by postnatal day 45,and the enriched stratum spinosum cells play crucial roles in VFA energy metabolism and immune capacity.Importantly,we identified the growth factorTBX3as a potential marker gene for papillary growth by interacting withPI3,thus highlighting the need to validate the role ofTBX3in papillogenesisin vivoandin vitro.
Our findings provide cellular-level insights into the molecular characteristics of morphogenesis,notably keratinization and papilla formation,and functional establishment (metabolism and immunity) of the rumen.This study will contribute to our understanding of rumen development and the interactions between dietary metabolism and the rumen.
DATA AVAILABILITY
The single-cell RNA-sequencing data were deposited in the Genome Sequence Archive database (http://gsa.big.ac.cn/)under accession No.CRA007511,Science Data Bank(doi:10.577 60/sciencedb.j00139.00007),and NCBI under BioProjectID PRJNA857336.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Q.Q.,J.H.L.,Z.P.L.,and S.Y.M.designed the project and research;D.M.S.,J.H.L.,W.Y.Z.,Y.Y.Y.,and J.H.collected samples and constructed sequencing libraries;Y.Y.,D.M.S.,and T.Q.conducted single-cell data analysis;Q.Q.,J.H.L.,Z.P.L.,and Y.Y.wrote the manuscript.Q.Q.,J.H.L.,Z.P.L.,R.H.,and Y.Y.amended the manuscript.All authors read and approved the final version of the manuscript.
- Zoological Research的其它文章
- Zoological Research call for papers of Cavefish Special Issue
- Fuel source shift or cost reduction:Context-dependent adaptation strategies in closely related Neodon fuscus and Lasiopodomys brandtii against hypoxia
- Ecological study of cave nectar bats reveals low risk of direct transmission of bat viruses to humans
- Population and conservation status of a transboundary group of black snub-nosed monkeys (Rhinopithecus strykeri) between China and Myanmar
- Nucleus accumbens-linked executive control networks mediating reversal learning in tree shrew brain
- Europe vs.China:Pholcus (Araneae,Pholcidae) from Yanshan-Taihang Mountains confirms uneven distribution of spiders in Eurasia

