Prior exposure to ciprofloxacin disrupts intestinal homeostasis and predisposes ayu (Plecoglossus altivelis) to subsequent Pseudomonas plecoglossicidainduced infection
Xiang-Yu Wu ,Jin-Bo Xiong ,Chen-Jie Fei ,Ting Dai ,Ting-Fang Zhu ,Zi-Yue Zhao ,Jing Pan,Li Nie,*,Jiong Chen,*
1 State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Ningbo University, Ningbo,Zhejiang 315211, China
2 Laboratory of Biochemistry and Molecular Biology, School of Marine Sciences, Meishan Campus, Ningbo University, Ningbo, Zhejiang 315211, China
3 Collaborative Innovation Center for Zhejiang Marine High-Efficiency and Healthy Aquaculture, School of Marine Sciences, Ningbo University, Ningbo, Zhejiang 315211, China
ABSTRACT With the rapid development of intensive farming,the aquaculture industry uses a great many antibiotics for the prevention and treatment of bacterial diseases.Despite their therapeutic functions,the overuse and accumulation of antibiotics also pose a threat to aquaculture organisms.In the present study,ayu (Plecoglossus altivelis) was used as a fish model to study the impacts of ciprofloxacin (CIP)overuse on intestinal homeostasis and immune response during subsequent Pseudomonas plecoglossicida infection.Based on 16S rRNA gene amplification and Illumina sequencing,we found that CIP pre-exposure caused significant variation in intestinal microbiota,including increased species richness,altered microbiota composition and interaction networks,and increased metabolic dysfunction.Furthermore,immunohistochemical analysis indicated that CIP pre-exposure resulted in severe mucosal layer damage,goblet cell reduction,and epithelial cell necrosis of the intestinal barrier in infected ayu.Quantitative real-time polymerase chain reaction (qRT-PCR) showed that disruption of intestinal homeostasis impaired systemic antiinfection immune responses in the intestine,gill,spleen,and head kidney,while inhibiting IL-1β,TNFα,and IL-10 expression and promoting TGF-β expression.Our findings indicated that CIP administration can directly affect intestinal microbiota composition and intestinal integrity in ayu fish.This perturbation of intestinal homeostasis is likely responsible for the lower survival rate of hosts following subsequent infection as the capacity to mount an effective immune response is compromised.This study also provides preliminary clues for understanding the effects of antibiotic overuse on higher vertebrates through trophic transfer.
Keywords:Ciprofloxacin;Microbiota;Intestinal barrier;Immune responses;Ayu
INTRODUCTION
With the rapid development of intensive aquaculture,outbreaks of various bacterial diseases continue to limit fishery productivity,resulting in losses of 50–100 billion yuan every year in China alone (Wang et al.,2015).As such,fish farmers use a great many antibiotics to prevent and treat these diseases,although often unnecessarily or inappropriately.At present,a total of 32 different antibiotics are used in aquaculture,mainly via oral administration (Liu et al.,2017).As a result,antibiotic residues have been detected in many aquatic products,with ciprofloxacin (CIP),norfloxacin,and sulfisoxazole accounting for the highest concentrations in China (Liu et al.,2017).CIP is a second-generation fluoroquinolone antibiotic used to treat aerobic gram-negative and gram-positive bacterial infections in humans,animals,and aquaculture species (Ebert et al.,2011;Nie et al.,2017).However,farmed aquaculture species are always exposed to higher than environmentally relevant concentrations of CIP.First,although farm ponds contain a large body of water,there is less variation compared to wild conditions,and CIP is more likely to accumulate but less biodegradable.Second,for effective disease prevention and treatment in aquaculture practice,CIP is typically used at concentrations of 1–50 mg/L(Liu et al.,2017;Nouws et al.,1988;Tran et al.,2018).Third,aquatic organisms cannot metabolize antibiotics efficiently,with 75% of antibiotics released back into the water in an unused form (Bhavani et al.,2022).Furthermore,although not all countries have approved CIP for animal use,it is a metabolite of the veterinary approved antibiotic enrofloxacin and is poorly biodegradable (Girardi et al.,2011;Kümmerer et al.,2000),resulting in higher concentrations of CIP in water,increased antimicrobial resistance,and restricted development of the aquaculture industry.
The fish intestine is host to many microbes and a primary target tissue of CIP in water.As an immune tissue,the intestine plays an essential role in maintaining homeostasis and initiating immune responses against enteric pathogens.After colonization,the intestinal microbiota provides nutrients to the host through fermentation end-products and secretory products.In addition,the microbiota inhibits the invasion of aquaculture pathogens by activating innate and adaptive immune cells.Although the fish microbiome contains many complex bacterial species,not all taxa contribute equally to the microbial community during pathogen invasion (Lu et al.,2020;Nie et al.,2017;Xiong et al.,2019).Using network analysis,recent studies identified candidate taxa that directly antagonize pathogens or provide favorable conditions for their growth during disease progression (Huang et al.,2020).
In addition to the intestinal microbiota itself,interactions between commensal and intestinal epithelial cells (IECs) are also critical for the maturation and function of the fish immune system and host homeostasis (Andriyanto et al.,2022;Rooks&Garrett,2016;Thoo et al.,2019).Fish IECs constitute the intestinal epithelial barrier and serve as the primary defense against intestinal invasion by pathogens (Wang et al.,2018).IECs interact with a vast array of intestinal microbes as well as innate and adaptive immune cells to maintain homeostatic conditions (Soderholm &Pedicord,2019).Conversely,intestinal microbes regulate IEC growth and function,including mucus secretion,tight junction integrity,and differentiation(Wang et al.,2018).Crosstalk between intestinal microbes,IECs,and systemic immune responses maintains fish homeostasis and inhibits pathogen colonization.
Studies in higher vertebrates have reported significant associations between antibiotic exposure and subsequent risk of infection (Malik et al.,2018;Li et al.,2020b).For example,recent antibiotic use can lead to subsequentClostridium difficile-associated infection due to disruption of the intestinal microbial community (Chang et al.,2008).Prior antibiotic exposure through commercial chicken is a risk factor forCampylobacter jejuniinfection (Effler et al.,2001).In human patients,exposure to antibiotics is associated with further risk ofSalmonella(Gradel et al.,2008;Neal et al.,1994;Pavia et al.,1990),Haemophilus influenzae(McVernon et al.,2008),andStaphylococcus aureusinfections (Early &Seifried,2012).Antibiotic use is also associated with increased fungal infections (MacDonald et al.,1993;Spinillo et al.,1999;Xu et al.,2008).Furthermore,antibiotic exposure and accumulation may significantly affect intestinal microbial composition,thereby impairing the metabolism and production of functional metabolites,providing vacant niches for pathogenic bacteria to breach the IEC barrier,and compromising anti-infection immune responses (Duan et al.,2022;Hagan et al.,2019;Zimmermann &Curtis,2019).However,despite the excessive application of CIP in the aquaculture industry,its association with subsequent immune responses,especially disruption of intestinal microbes,remains poorly studied in aquatic organisms.
In addition to the direct effects of CIP overuse on aquatic organisms,long-term antibiotic exposure may also promote the occurrence and spread of antibiotic-resistant bacteria(ARB) and antibiotic-resistance genes (ARGs) (Guan et al.,2018;Jiang et al.,2013;Xue et al.,2021).Notably,CIP in water can accumulate in multiple tissues and organs of higher trophic fish,with subsequent transfer to terrestrial organisms,including humans,via predator-prey relationships (He et al.,2016;Zhang et al.,2021;Zhao et al.,2021).Therefore,studying the accumulation of CIP in the intestinal microbiota and consequent anti-pathogenic immune response is critical,not only for the aquaculture industry,but also for the long-term health of humans.
In the present study,we explored the effects of CIP exposure on intestinal microbiota homeostasis,intestinal barrier structure,and subsequent anti-infection immune responses in ayu (Plecoglossus altivelis).This study should expand our understanding of the detrimental effects of antibiotic overuse on aquaculture organisms,especially subsequent pathogen infection,and should provide insights regarding the transfer of these harmful effects to higher vertebrates through predator-prey relationships.
MATERIALS AND METHODS
Fish rearing
A total of 84 ayu (approximately 50 g) were purchased from a commercial fish farm in Xiangshan County,Ningbo,China.The fish were randomly dispersed into 14 tanks (60 L capacity,six fish per tank) and maintained in a recirculating water system (21.0±1.0 °C) with regular feeding.All applicable international and national guidelines for the care and use of animals were strictly followed.All experiments involving animals were approved by the Ningbo University Institutional Animal Care and Use Committee and were carried out in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Antibiotic and pathogen administration and sample collection
ThePseudomonas plecoglossicida(NZBD9) bacteria were a kind gift from Professor Yan’s Lab (Huang et al.,2018) and were cultured in Luria Bertani (LB) medium at 18 °C for 12 h with shaking at 220 r/min.In total,12 fish were administered CIP (final concentration 1 mg/L) and 18 fish were treated with phosphate-buffered saline (PBS) (control blank group,CK).After treating the fish with PBS or CIP for 7 d,antibiotic exposure was ceased and six fish from each group were sampled (designated as CK pre-infection (CK pre) and CIP pre-infection (CIP pre),respectively).The remaining fish in the PBS and CIP-treated groups were challenged withP.plecoglossicidafor 7 d (final concentration 1×106CFU/mL)through bath immersion (designated asP.ppostinfection (P.p.post) and CIP+P.ppostinfection (CIP+P.ppost),respectively).The PBS-treated group was set as the control (CK post,Figure 1).Fish from each group were sacrificed and immune tissues,including intestine,gill,spleen,liver,and head kidney,were collected at indicated time points for RNA isolation:CK pre,CIP pre,CK post,P.ppost,and CIP+P.ppost (Figure 1).Intestine samples of each group were aliquoted in three parts,one for genomic DNA extraction,one for total RNA isolation,and one for histological analysis.
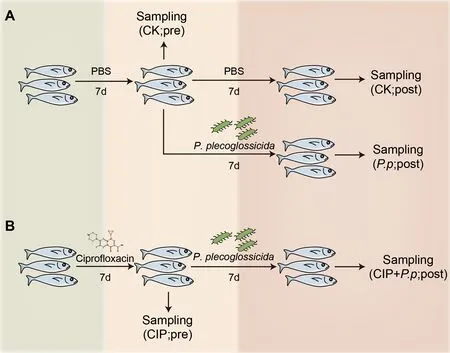
Figure 1 Schematic showing specific procedures and sampling times of each group
The remaining 54 fish were divided into three groups.One group was exposed to CIP (1 mg/L) and the other two groups were treated with PBS alone for the first 7 d.After 7 d,CIP exposure was ceased and the fish were further infected withP.plecoglossicida,with one PBS-treated group set as the control.The different treatment groups were designated as CK(PBS treated),P.p(P.plecoglossicidainfected),and CIP+P.p(CIP pretreated andP.plecoglossicidainfected).The survival rate of each group was calculated on day 14 following antibiotic exposure.
16S rRNA gene amplification and Illumina sequencing
Genomic DNA was extracted according to the manufacturer’s guidelines (Qiagen,USA).The bacterial V3–V4 region of the 16S rRNA gene was amplified from extracted genomic DNA using the primer pair 341F (5′-CCTAYGGGRBGCASCAG-3′)and 806R (5′-GGACTACNNGGGTATCTAAT-3′).Polymerase chain reaction (PCR) was performed under the following conditions:95 °C for 2 min,25 cycles of denaturation at 95 °C for 30 s,annealing at 55 °C for 30 s,and extension at 72 °C for 45 s,with a final extension at 72 °C for 10 min.Triplicate amplicons for each sample were pooled and purified using a PCR fragment purification kit.The concentration of purified amplicons was measured using a PicoGreen-iT dsDNA Assay Kit (Invitrogen,USA).The same amount of amplicons from each sample were pooled and sequenced using the NovaSeq 6000 platform (Illumina,USA).
Processing and analysis of Illumina sequencing data
Fastp (v0.14.1,https://github.com/OpenGene/fastp) was used to control raw data quality via the sliding window method(Option:-W 4 -M 20) to obtain paired-end clean tags.The tags were merged using FLASH (Magoč &Salzberg,2011).The filtered data were assembled using the QIIME2 pipeline(Bolyen et al.,2019).Sequences with more than three truncated or ambiguous consecutive bases at any site with a Phred quality score Q<25 were excluded.Chimeras were identified and removed using the UCHIME algorithm (Edgar et al.,2011).Bacterial phylotypes were identified using the UCLUST algorithm and classified into zero-radius operational taxonomic units (zOTUs) at a 97% similarity threshold (Edgar,2010).The most abundant sequence of each zOTU was selected as its representative sequence and taxonomically assigned against the Greengenes database (v13.8) using PyNAST (Caporaso et al.,2010).Taxonomic information was annotated using the RDP Classifier (v2.2,http://sourceforge.net/projects/rdp-classifier/) with a 90% confidence threshold(Wang et al.,2007).After taxonomies were annotated,archaea,eukaryotes,chloroplasts,and those unassigned to the bacterial domain were excluded from the zOTU table.To avoid sequencing effort bias,we used a randomly rarefied subset of 40 000 sequences per sample in subsequent analysis (excluding one sample with a sequence/number of reads ratio <40 000).
Microbial co-occurrence network construction
To determine gut microbial interactions in response to perturbations (CIP exposure andP.plecoglossicidainfection),microbial co-occurrence networks were constructed using the igraph package in R (Csardi &Nepusz,2006) based on zOTU absolute abundance.Those zOTUs with an abundance >0.5%of the rarefied count matrix were selected.Random matrix theory (RMT) was used to automatically identify the appropriate similarity threshold (St) before network construction,and all networks were constructed using the same St (Shi et al.,2016).Correlation between two zOTUs was considered strong if the Spearman correlation coefficient(ρ) was >0.7 and statistically significant (adjustedP<0.001;Benjamini-Hochberg) (Weiss et al.,2016).Covariations were detected with six biological replicates for each network,and only zOTUs detected in more than half of the samples of each group were retained for network construction.Statistical analysis was performed using the vegan,igraph,and Hmisc packages in R (Csardi &Nepusz,2006;Harrell &Harrell,2019;Oksanen et al.,2013) and networks were visualized using Gephi (v0.9.0) (Bastian et al.,2009).
The co-occurrence network data were further screened for zOTU members directly associated withP.plecoglossicida.Only interactions includingP.plecoglossicidawere retained for further analysis.Networks based on these correlations were visualized using Cytoscape (v3.3.0) (Shannon et al.,2003).All parameters were the same as described above.
Functional profiles of intestinal microbiota
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) analysis was used to predict functional genes based on the 16S rRNA gene composition of different groups (Langille et al.,2013).The zOTU tables were rearranged using the QIIME2 closed reference plugin with 97% similarity to the database (Bolyen et al.,2019) and imported into PICRUSt.The classified functional genes (Kyoto Encyclopedia of Genes and Genomes (KEGG)) were further analyzed according to the developer’s guidelines using the Galaxy platform (http://huttenhower.org/galaxy) (Jalili et al.,2020).Statistical analysis and visualization were performed using STAMP (v2.1.3).
Intestinal histology
Histological analysis was performed on intestinal tissue collected from each group at relevant time points.The samples were fixed with 10% formaldehyde for 24 h at room temperature,dehydrated with an ethanol gradient,transparentized with xylene,and embedded in paraffin wax.The intestinal paraffin blocks were cut into sections (3 μm),mounted on slides,deparaffinized and rehydrated,and finally stained with hematoxylin &eosin (H&E).The slides were subsequently observed under a microscope (Eclipse Ci-L,Nikon,Japan).
Quantitative real-time PCR (qRT-PCR)
Tissue total RNA was isolated using RNAiso Plus (Takara,China),and reverse-transcribed into first-strand cDNA using a reverse transcription kit (Takara) according to the manufacturer’s instructions (Li et al.,2020a;Shen et al.,2020).The expression levels ofpaIL-β,paTNF-α,paTGF-β,andpaIL-10were analyzed by qRT-PCR using primers listed in Supplementary Table S1 with SYBR Premix Ex Taq II(Takara Bio) on an ABI StepOneTMReal-Time PCR System(Applied Biosystems,USA).Relative gene expression was calculated using the 2−ΔΔCTmethod and normalized againstpa18SrRNA(Wang et al.,2021).Each PCR trial was performed in triplicate and repeated at least three times.
Statistical analysis
The zOTU abundance data were normalized to the standard sample with the fewest sequences.Subsequent analyses of alpha and beta diversities were performed based on the normalized data.The alpha diversity index,i.e.,diversity within the samples,was calculated using QIIME2,and included the Chao1 and richness indices for species richness and the Shannon and Simpson indices for species evenness (Thukral,2017).Data were plotted using GraphPad (v8.1.0) and significance of alpha diversity was determined using the paired two-sided Wilcoxon test in R (v3.6.0).To analyze beta diversity,principal coordinates analysis (PCoA) and analysis of similarity (ANOSIM) were performed with the Bray-Curtis metric using the vegan package in R (v3.6.0).Permutational multivariate analysis of variance (PERMANOVA) was used to assess the impacts of CIP andP.plecoglossicidaon variations in intestinal microbiota composition of ayu with the Adonis function in the vegan package of R (Oksanen et al.,2013).
RESULTS
Effects of CIP exposure on survival and pathogen abundance of P.plecoglossicida-infected ayu
The survival rates of ayu administered CIP andP.plecoglossicidaalone or in combination were determined to evaluate the role of CIP exposure on host defense against pathogens (Figures 1,2A).Daily monitoring results showed that CIP pretreatment had no obvious life-threatening effects on ayu.However,when further infected withP.plecoglossicida,the survival rate of the CIP-pretreated group(CIP+P.p) dramatically decreased to 23.86% at day 14,while the survival rate of theP.plecoglossicida-only infected group(P.p) remained at 67.13% (log-rank (Mantel-Cox) test,*:P<0.05).Further investigation revealed that the CIP+P.pgroup had the highest intestinal abundance ofP.plecoglossicida(zOTU9),consistent with the survival results(Figure 2B).In summary,CIP administration promoted introduced pathogen proliferation and increased host susceptibility,most likely by altering intestinal microbial composition.
CIP exposure altered alpha diversity of intestinal microbiota in P.plecoglossicida-infected ayu
To verify whether the effects of CIP exposure on intestinalP.plecoglossicidaabundance was due to alteration in intestinal microbial composition,16S rRNA gene amplicon sequencing was conducted to capture bacterial community composition.Optimized reads (ranging from 29 619 to 80 639) were obtained from all samples.Samples with less than 40 000 reads were excluded from analysis.
Alpha diversity (species richness and evenness) was analyzed for the zOTU data to evaluate overall taxonomic diversity between groups.CIP pretreatment had no obvious effects on microbial alpha diversity (Figure 3A,CK pre and CIP pre).However,when subsequently administeredP.plecoglossicida,CIP pretreatment significantly increased taxonomic diversity (CIP+P.p),as measured by the Chao1 and richness indices (Figure 3A,*:P<0.05).TheP.plecoglossicida-only-infected group (P.p) was characterized by a lower Chao1 index,indicating that pathogen infection may disrupt the balance of healthy intestinal microbiota(Figure 3A,*:P<0.05).Species evenness was comparable between groups (Figure 3B).
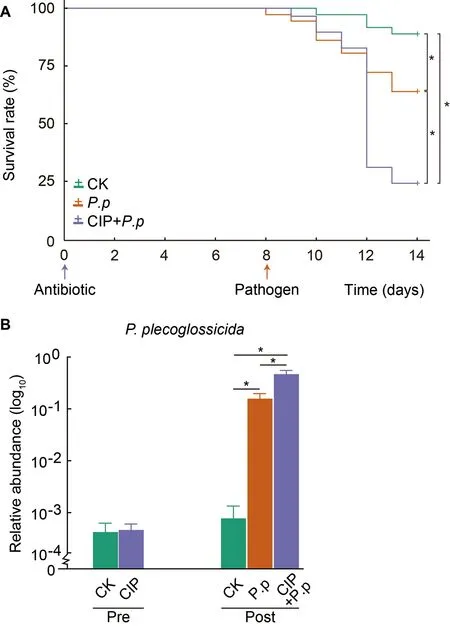
Figure 2 CIP exposure affects survival and pathogen abundance of P.plecoglossicida-infected ayu
CIP exposure altered intestinal microbial communities in P.plecoglossicida-infected ayu
PCoA was conducted to evaluate the effects of CIP on intestinal microbial community composition in theP.plecoglossicida-infected ayu.Before infection,the CIPexposed group (CIP) community was clustered far from that of the control group (CK) (PERMANOVA using Bray-Curtis metric,P=0.002,Figure 4A,top left),revealing that CIP had a significant impact on the composition of intestinal microbiota.In addition,P.plecoglossicidainfection (P.p) altered the intestinal microbial community (P=0.004,Figure 4A,top right),but no significant difference was observed when coadministered with CIP (CIP+P.p) compared with the control group (CK,post) (P=0.079,Figure 4A,bottom left).The microbial composition of the CIP+P.pgroup also significantly differed from that of theP.pgroup (P=0.002,Figure 4A,bottom right).
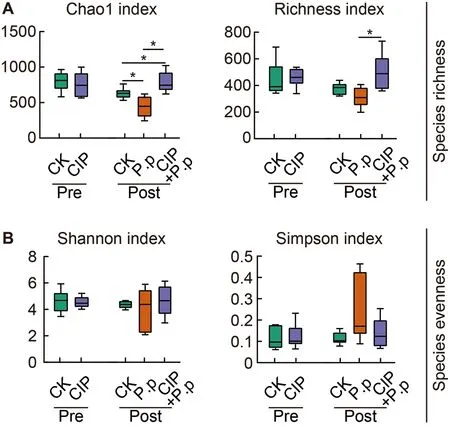
Figure 3 Effects of CIP exposure on alpha diversity of P.plecoglossicida-infected ayu microbiota
The relative abundances of dominant bacteria were further analyzed at the phylum level to explore changes in bacterial composition in the fish intestine under different conditions(Figure 4B).Results showed that bacterial composition remained essentially consistent in the CK groups over time(pre vs.post infection),withProteobacteria(25.7%),Bacteroidetes(22.7%),Firmicute(23.3%),Actinobacteria(20.9%),andTenericutes(10.4%) identified as the most dominant phyla.FollowingP.plecoglossicidainfection,Actinobacteriaincreased markedly with a concomitant reduction in other dominant phyla.CIP exposure increased the prevalence ofProteobacteriaandFirmicutesto 42.6% and 48.1%,respectively,accompanied by a decrease in the abundances ofActinobacteria,Bacteroidetes,andTenericutes.Antibiotic exposure had enduring effects on intestinal microbiota,as very similar bacterial compositions were observed in the CIP-exposed andP.plecoglossicidainfected (CIP+P.p) group compared to fish exposed to antibiotics alone (CIP group).
When representative bacteria were classified into genera,distinct differences emerged between fish in the CIP-exposed groups (CIP and CIP+P.p) versus the control (CK) andP.plecoglossicida-infected groups (P.p).Notably,Prevotella_9,the main genus in the phylumBacteroidetes,was rare in the CIP-exposed groups (CIP and CIP+P.p) compared with the other groups (Figure 4C).Several zOTUs were dominant in the CIP-treated fish.For instance,the relative abundances ofAncylobacterandStreptococcuswere elevated after antibiotic exposure with or without infection.These taxa could be identified as bacterial indicators for distinct groups.
CIP exposure altered co-occurrence networks of intestinal microbiota of P.plecoglossicida-infected ayu
To assess changes in bacterial ecosystem structure induced by CIP exposure duringP.plecoglossicidainfection,the cooccurrence patterns of microbial communities at the zOTU level were determined based on Spearman correlations(Figure 5).Only strong connections (Spearman correlation coefficientsρ>0.7) between zOTUs with a relative abundance above 0.5% were selected.A slight decrease in node number and 2-fold decrease in edge number were observed in the CIP-exposed group before infection (Table 1).When coadministered withP.plecoglossicidainfection,CIP exposure(CIP+P.p) compensated for the effects of pathogen infection by reducing the number of nodes and edges (2-and 4-fold,respectively) compared to theP.pgroup.
Most associations (edges) were positive for all networks and mean degree was the best predictor across all network projection types (Banerjee et al.,2019).AfterP.plecoglossicidainfection,the mean degree distribution in theP.pnetwork was much higher than that in the CIP-pretreated network (CIP+P.p)(Table 1).In addition,betweenness centrality and degree of centralization strength of the network decreased,indicating that CIP pretreatment induced a relatively decentralized co-occurrence network of the intestinal microbiota compared with the pathogen-only-infected group.
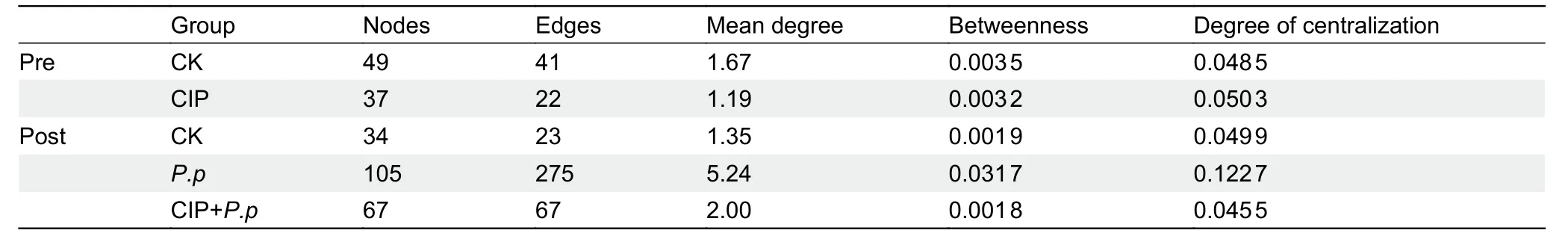
Table 1 Co-occurring bacterial OTU network metrics based on Spearman correlation analysis of ayu gut microbiota for overall groups
Four major phyla (Proteobacteria,Firmicutes,Bacteroidetes,andActinobacteria) were included in all cooccurrence networks of the different groups (Figure 5).Prior to infection,Bacteroidetescontributed the largest number of nodes,followed byActinobacteria(Figure 5A,C,CK pre).CIP treatment substantially increased the abundance and centralization of theFirmicutesandActinobacterianodes,while subclusters of zOTUs belonging toBacteroidetesalmost disappeared (Figure 5A,C,CIP pre).After infection,theP.pgroup network displayed marked changes,with more varied species abundance and more intense subcluster formations compared with the CIP+P.pnetwork (Figure 5B,C).TheFirmicutesandBacteroidetesphyla were highly interconnected,with the most interactions in theP.pnetwork.Compared with theP.pgroup,the CIP+P.pgroup exhibited more negative correlations within OTUs and the formation of peripheral bacterial clusters.The CIP+P.pnetwork also presented more subclusters of OTUs belonging toProteobacteriacompared to theP.pgroup.
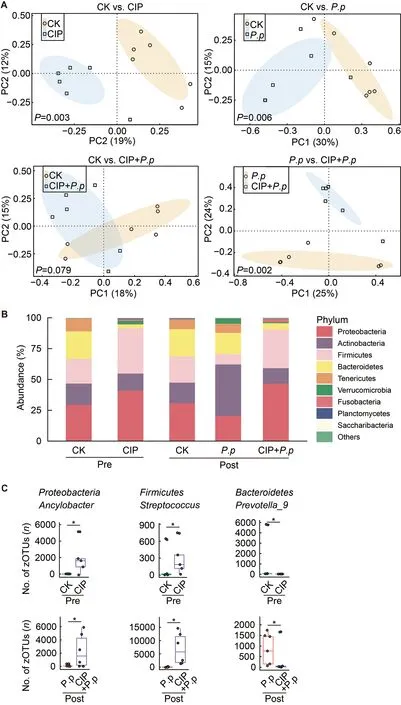
Figure 4 Effects of CIP exposure on intestinal microbial community in P.plecoglossicida-infected ayu
As CIP exposure had a significant influence on the overall co-occurrence networks of the microbial community,the taxa directly related toP.plecoglossicidawere further analyzed.We focused on theP.plecoglossicidamodules to identify taxa that promote or prevent infection.Results indicated that the topologies of theP.plecoglossicidanetwork interactions changed significantly after CIP exposure or pathogen infection.When exposed to CIP,taxa related to backgroundP.plecoglossicidaincreased,withEnterococcus(zOTU61),Streptococcus(zOTU99),andAeromonas(zOTU108)showing positive interactions andAncylobacter(zOTU17),Alpinimonas(zOTU20),andBacteroides(zOTU87) showing negative interactions (Figure 6A).When further infected withP.plecoglossicida,the networks became more complicated and the prevalence of interacting zOTUs increased(Figure 6B).A special co-occurrence pattern was observed in theP.pgroup,with 10 negative interactions and one positive interaction.The high negative/positive interaction ratio highlighted the protective role of the host microbiota against pathogen invasion.However,the interacting taxa differed markedly when fish were pretreated with CIP before infection(CIP+P.p),with the number of “contributors” increasing,number of “inhibitors” decreasing,and negative/positive ratio decreasing to 1:1.Mycoplasmataceae(zOTU13),Ancylobacter(zOTU17),Vibrio(zOTU33),Bacterium_TSA331-4(zOTU148),Lachnospiraceae(zOTU182),Ruminiclostridium_9(zOTU261),andRuminococcus_1(zOTU303) showed positive interactions with the pathogen.Of note,Ancylobacter(zOTU17) in the CIP-administered group (CIP,Figure 6A) hindered backgroundP.plecoglossicidacolonization,indicating that interactions between two taxa may alter upon environmental change.Prevotella(zOTU328,zOTU442),GKS98_freshwater(zOTU475),Ruminococcaceae(zOTU558),Bacteroidales_S24-7(zOTU639),Clostridium_sp._ID11(zOTU705),andRuminococcaceae(zOTU774) were antagonized byP.plecoglossicidaduring infection (Figure 6B).
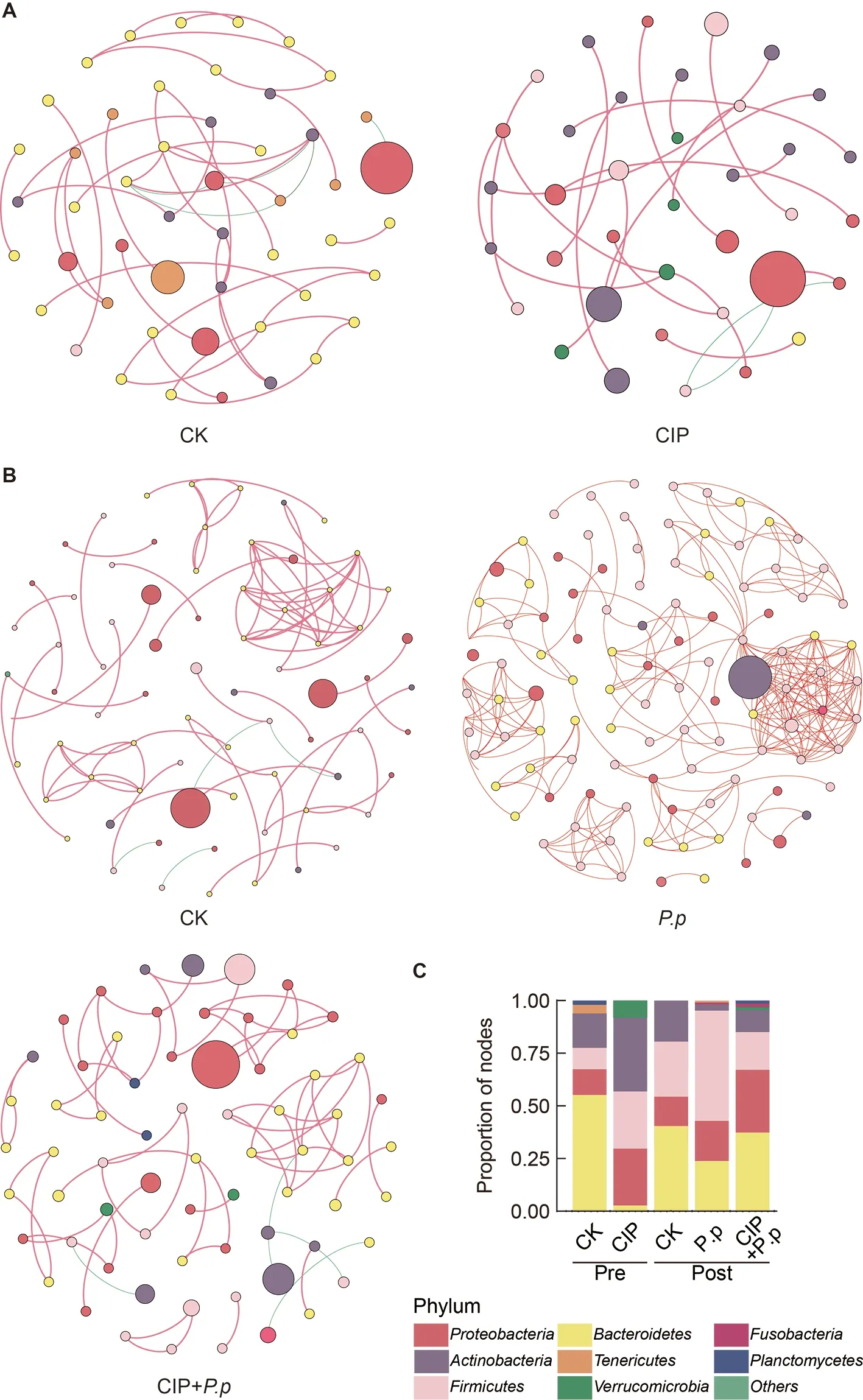
Figure 5 Effects of CIP exposure on the co-occurrence patterns of the intestinal microbiota in P.plecoglossicida-infected ayu
CIP exposure altered functional genes of intestinal microbiota
PICRUSt was applied to explore functional gene profiles of the intestinal microbiota in different groups.Pathways associated with amino acid metabolism were significantly enhanced when ayu were pretreated with CIP (CIP vs.CK,Figure 7A,P<0.05).When infected withP.plecoglossicida,energy metabolismrelated pathways were enriched but carbohydrate metabolismrelated pathways were repressed (Figure 7B,P.pvs.CK,P<0.05).For CIP pretreatment beforeP.plecoglossicidainfection (CIP+P.p),pathways involved in cofactors and vitamins decreased compared with the CK andP.pgroups,and pathways related to nucleotide metabolism and cofactors and vitamins also decreased compared to the control(CIP+P.pvs.CK) andP.plecoglossicida-infected (CIP+P.pvs.P.p) groups,respectively (Figure 7B,P<0.05).
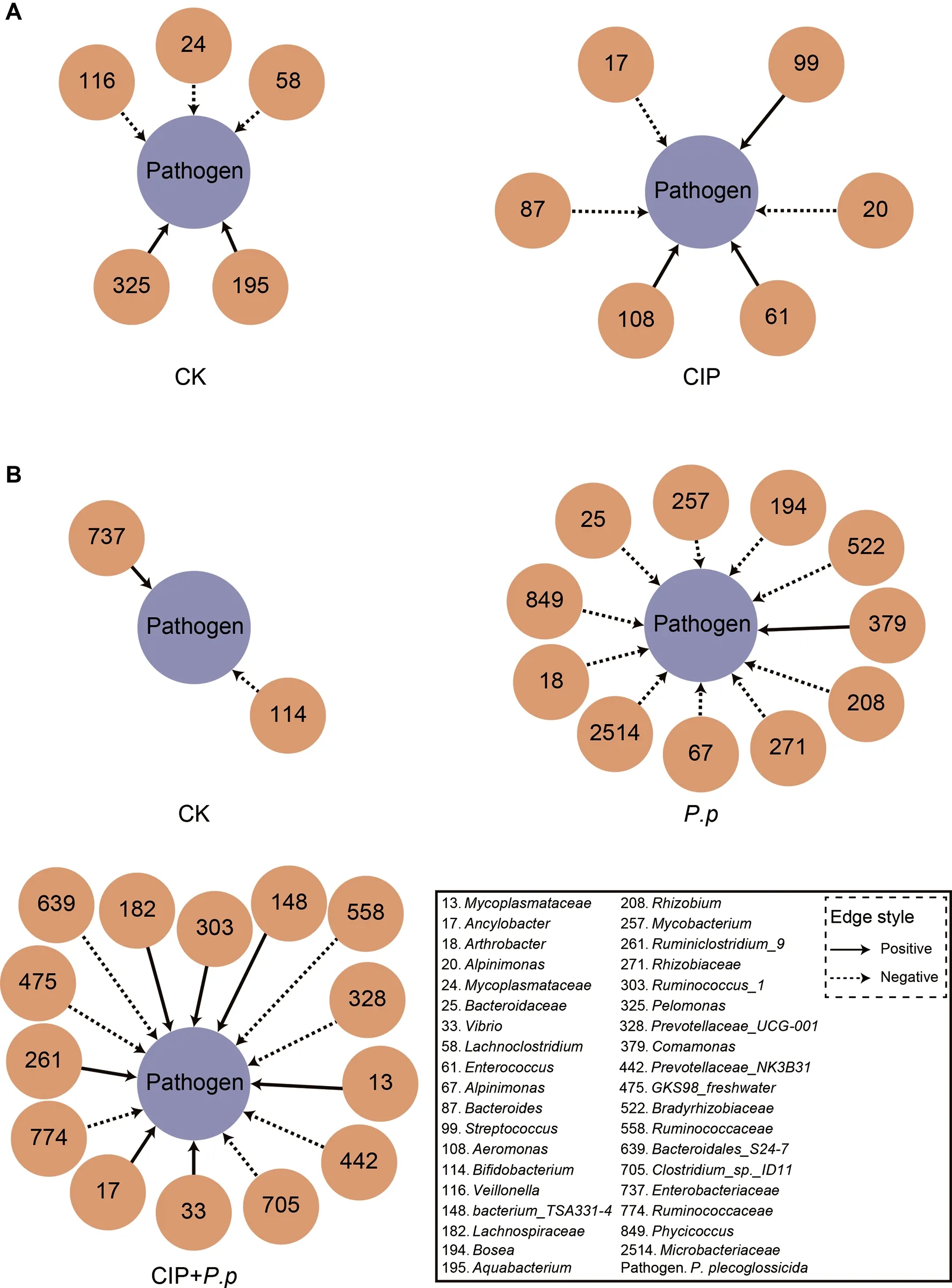
Figure 6 Interspecies interactions of pathogen-host taxa in ayu intestinal microbiota
CIP exposure affects intestinal epithelial barrier structure in P.plecoglossicida-infected ayu
As interactions between the intestinal epithelial barrier and microbes are important for host homeostasis,we investigated whether the CIP-induced changes in microbial biodiversity also affect the structure of the intestinal epithelial barrier.Based on H&E staining,compared to the regularly arrayed intestinal villi with normal height and width and tightly arrayed epithelial cells in the CK group (Figure 8A,left),the intestinal epithelial cells in the CIP-treated group showed severe necrosis and a considerable decrease in goblet cells(Figure 8A,right,and F),indicating that CIP accumulation destroyed the normal structure of the intestinal epithelial barrier.AfterP.plecoglossicidainfection,villus height decreased,villus width increased,and goblet cell number increased markedly compared with the CK group (Figure 8B,upper right,and F).CIP pretreatment further aggravatedP.plecoglossicida-induced intestinal epithelial barrier damage,characterized by complete mucosal layer destruction,epithelial cell necrosis,and goblet cell reduction (Figure 8B,lower left,and F).
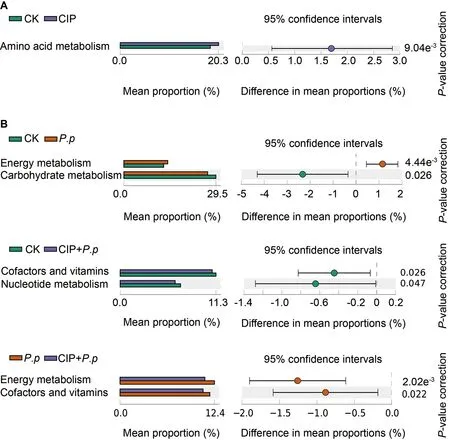
Figure 7 Predicted functions of intestinal microbiota of ayu after CIP exposure and P.plecoglossicida infection
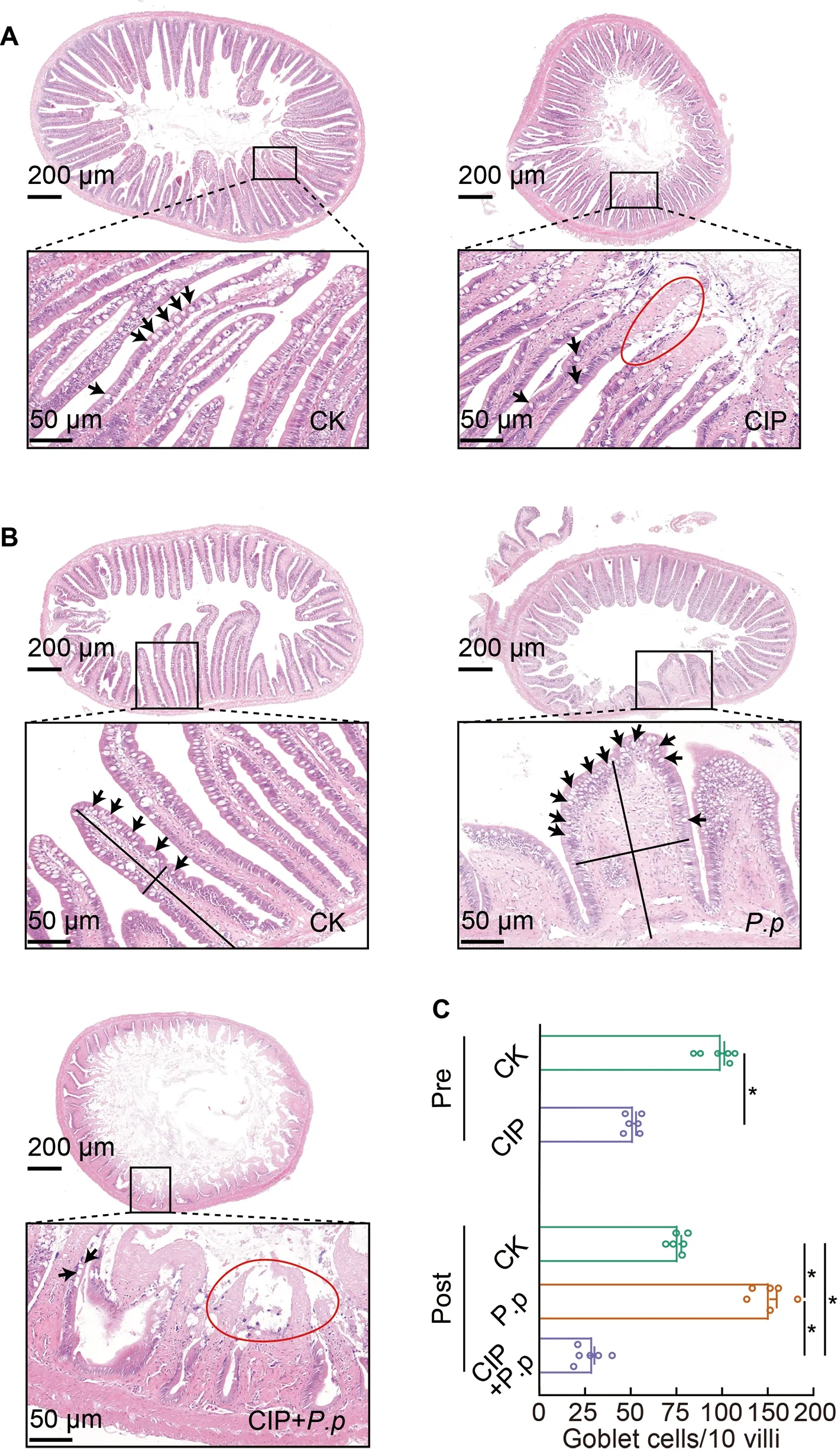
Figure 8 Intestinal histological changes in ayu after CIP and P.plecoglossicida administration
CIP exposure shapes immune response in P.plecoglossicida-infected ayu
To further investigate the effects of CIP exposure on the systemic cytokine profiles of ayu in response toP.plecoglossicidainfection,qRT-PCR analysis of canonical proand anti-inflammatory cytokines in the intestine (Figure 9A),gill (Figure 9B),spleen (Figure 9C),and head kidney(Figure 9D) was performed.Results showed that CIP exposure (CIP group) significantly down-regulated proinflammatory cytokines (IL-1βandTNF-α) in most examined tissues,except forTNF-αexpression in the gill (unchanged)and spleen (significantly increased).FollowingP.plecoglossicidainfection (P.pgroup),the pro-inflammatory cytokine expression levels increased significantly in the spleen and head kidney,althoughTNF-αlevels remained comparable to the control.A 5-fold increase inIL-1βexpression was detected in the gill,while both pro-inflammatory cytokines showed a significant decrease in the intestine followingP.plecoglossicidainfection.In the CIP+P.pgroup,the proinflammatory cytokine profiles were further modified due to antibiotic exposure prior to infection.Fish pre-exposed to CIP were less responsive toP.plecoglossicidainfection(CIP+P.p),as the observed cytokine profiles were very similar to those of the CIP group.Compared to theP.pgroup,theIL-1βandTNF-αexpression levels were significantly reduced in most tissues,except for the gill and spleen,whereTNF-αexpression was significantly increased.In comparison,the two anti-inflammatory cytokines (TGF-βandIL-10) exhibited different expression profiles under these same conditions.Specifically,CIP exposure produced the opposite effect on cytokine profiles.IL-10expression was significantly reduced in all tissues,whileTGF-βexpression was significantly increased in the intestine,gill,and head kidney,but significantly decreased in the spleen.In theP.plecoglossicida-infected fish,although a slight but significant increase (1.2-fold) inIL-10expression was observed in the intestine and gill,P.plecoglossicidainfection primarily suppressed the expression of anti-inflammatory cytokines,withTGF-βandIL-10significantly reduced in most tissues.However,exposing fish to CIP prior to infection reversed the trends observed in theP.pgroup.For example,TGF-βexpression was significantly increased in the intestine,gill,and head kidney of the CIP+P.pgroup,whileIL-10expression was significantly reduced in the intestine and gill.The only exception occurred in the spleen,in whichTGF-βandIL-10expression levels were further decreased in the CIP+P.pgroup compared to theP.pgroup.
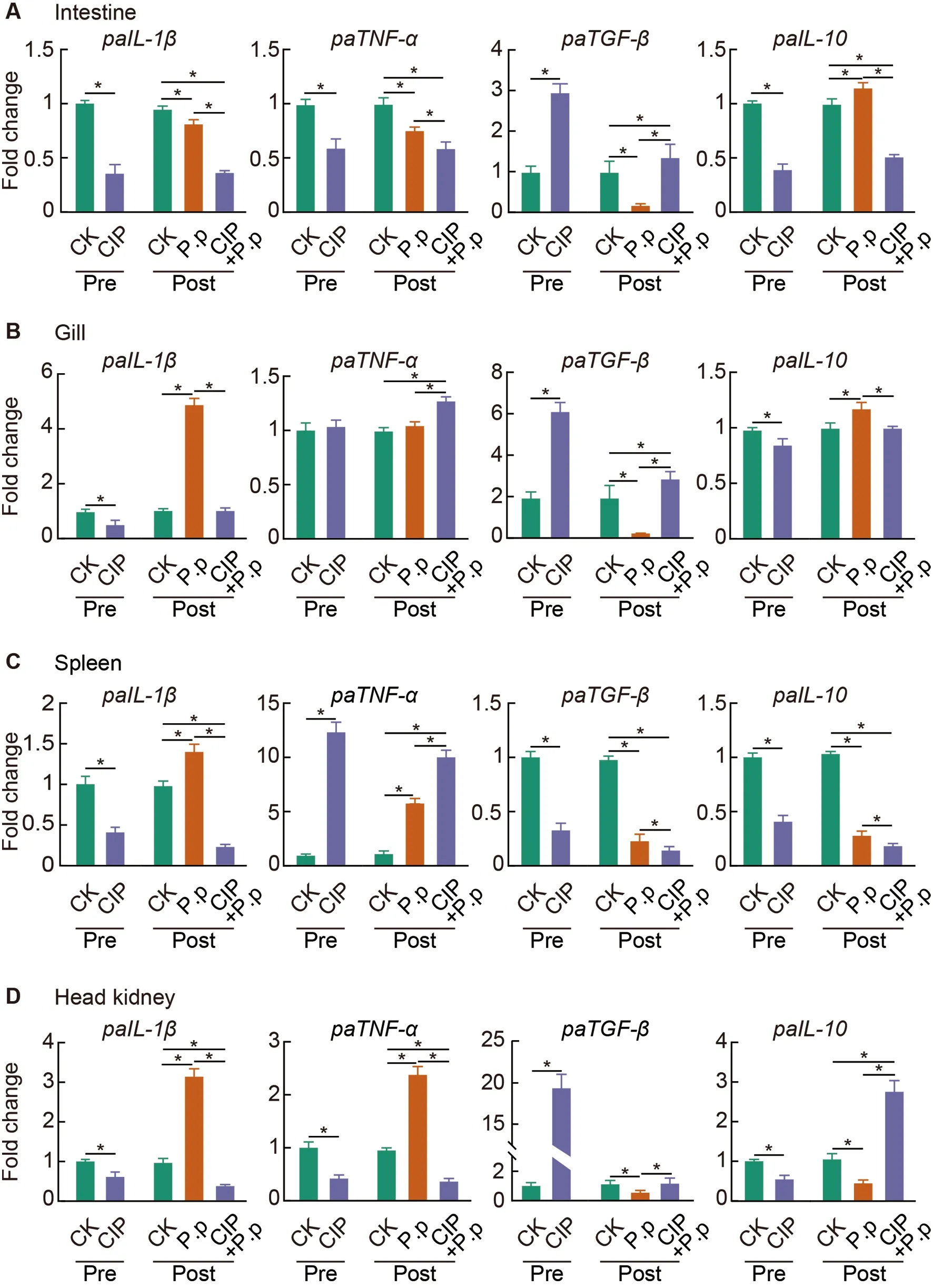
Figure 9 CIP exposure shapes immune responses of P.plecoglossicida-infected ayu
DISCUSSION
CIP is a type of fluoroquinolone antibiotic commonly used for the treatment of bacterial diseases in humans,animals,and aquaculture species.Intensive use and poor degradation have resulted in considerable CIP accumulation in the environment,particularly in aquatic ecosystems where very high doses of CIP are used for the prevention and treatment of diseases (Liu et al.,2017;Nouws et al.,1988;Tran et al.,2018).Fish farmers frequently administer high doses of antibiotics to prevent or limit unpredictable mortality,with CIP concentrations in fisheries of up to 50 mg/kg (50 mg/L equivalent) (Carlson et al.,2017;Liu et al.,2017).Although CIP exhibits potent antimicrobial activity to help control the spread of disease,it also exerts adverse impacts on aquatic species,e.g.,disruption of intestinal homeostasis as beneficial microbes are indiscriminately killed.In contrast to the adverse effects of CIP on mammals,its negative effects on fish and exact modes of action,especially on the risks of subsequent pathogen infection,remain largely unknown.Here,we used ayu as a model species to investigate the effects of high-dose CIP on intestinal homeostasis and the capacity of CIPexposed fish to mount an effective immune response during subsequentP.plecoglossicidainfection.The selected concentration of CIP (1 mg/L) was close to the lower limit used in aquaculture practice.Results showed that CIP exposure increased mortality in theP.plecoglossicida-infected ayu(Figure 2A),consistent with previous studies showing higher susceptibility to pathogen infection during antibiotic treatment(Roubaud-Baudron et al.,2019;Schubert et al.,2015;Sekirov et al.,2008).In addition,significantly enriched intestinalP.plecoglossicidawas observed in the CIP-pretreated andP.plecoglossicida-infected group (CIP+P.p) than in theP.plecoglossicida-only-infected group (P.p) (Figure 2B),partially explaining the higher observed mortality.
The effects of CIP pre-administration on pathogen-infected microbiota were evaluated.CIP exposure did not affect the overall taxonomic diversity of the intestinal microbiota (CIP),whileP.plecoglossicidainfection (P.p) slightly decreased species richness (Figure 3).However,when both stimulants were co-administered (CIP+P.p),species richness increased significantly and evenness remained unchanged (Figure 3),inconsistent with previous studies showing a significant decrease in taxonomic diversity in antibiotic-treated intestinal microbiota (Liu et al.,2017;Wang et al.,2019).Nevertheless,recent studies have also reported increased taxonomic diversity in mink fecal and Atlantic salmon intestinal microbiotas after exposure to antibiotics,with a general decrease in diversity indices after infection (Gupta et al.,2019;Marker et al.,2017).
Proteobacteria,Bacteroidetes,Firmicute,andActinobacteriaare the dominant phyla in fish intestinal microbiota (Wang et al.,2018).A similar bacterial composition was observed in the ayu intestine (Figure 4B).AfterP.plecoglossicidainfection,however,the percentages ofActinobacteriaandVerrucomicrobiaincreased,while the other dominant phyla declined (Figure 4B).Consistent with our observations,rats with intestinal dysbiosis and inflammatory disease are reported to show an increase inVerrucomicrobiaand a decrease inTenericutes(Lakshmanan et al.,2021).CIP exposure exhibited enduring effects on intestinal microbiota,as very similar bacterial compositions were observed in the co-administered (Figure 4B,CIP+P.ppost) and CIP-exposed groups (CIP pre),with a slightly higher prevalence ofProteobacteriaand lower prevalence ofVerrucomicrobiain the co-administered group.Prior research on Atlantic salmon and zebrafish has also indicated that antibiotic administration can shift intestinal microbial composition (Gupta et al.,2019;Kayani et al.,2021).Compared with the controls (CK pre and CK post),CIP exposure increased the prevalence ofProteobacteria,Firmicutes,andVerrucomicrobia,but decreased the abundance ofActinobacteria,Bacteroidetes,andTenericutes.Previous studies have indicated thatProteobacteriadominance disturbs intestinal microbiota and accelerates disease progression (Shin et al.,2015),and higherFirmicutes/Bacteroidetesratios are associated with intestinal dysbiosis in obesity models (Stojanov et al.,2020).This evidence is consistent with our ayu intestinal microbe results,indicating the detrimental role of CIP on ayu microbiota equilibrium.
At the genus level,higherAncylobacterandStreptococcuscounts were observed after CIP exposure (CIP and CIP+P.p,Figure 4C).Streptococcusaccommodates a wide range of gram-positive bacteria responsible for many fish diseases (El-Noby et al.,2021).As the dominant genus within the phylumBacteroidetes,Prevotellaincreased in theP.plecoglossicidainfected group compared to the co-administered group(Figure 4C).Prevotellaprevalence is associated with an increase in the release of inflammatory factors by various immune and stromal cells in the intestine,leading to excessive accumulation associated with immune disorders (Ilhan et al.,2020).Deficiency ofPrevotellain the CIP-exposed group may attenuate the immune response of pathogen-invaded ayu.
Antibiotic administration has been shown to reduce the number of nodes and edges in the cow foregut and hindgut microbial networks (Ji et al.,2018) and weaken the bacterial interaction networks of the chicken gut microbiota (Gao et al.,2017).In the present study,our results also showed that CIP treatment reshaped network structure in the ayu fish(Figure 5).Infectious diseases can alter microbial network features,forming more complex networks and more highly connected nodes compared to healthy subjects (Dai et al.,2020).Complex networks with core properties are more robust to external disturbance than simple networks (Santolini &Barabási,2018).With higher complexity and core nodes,theP.pnetwork exhibited higher resistance and protection against external invasion,while the less complex CIP+P.pnetwork showed higher susceptibility to pathogens (Figure 5B).
After CIP exposure,EnterococcusandStreptococcuswere defined as cooperative interspecies withP.plecoglossicida(Figure 6A).Consistently,EnterococcusandStreptococcusare considered as opportunistic pathogens in aquaculture(Krawczyk et al.,2021) and increasedStreptococcusis related to several farmed fish diseases (El-Noby et al.,2021).Pathogen invasion can substantially reshape homeostatic interactions within bacterial communities (Yang et al.,2017).Here,theP.pgroup contained more competitive members than the CIP+P.pgroup.The reduction in competitive species would promote pathogen growth after CIP exposure.In contrast,as cooperative taxa,Vibrio,Mycoplasmataceae,andAncylobacterexhibited a co-occurrence pattern in the CIP+P.pnetwork (Figure 6B).These cooperative species are generally considered opportunistic pathogens in aquaculture (Baker-Austin et al.,2018).
PICRUSt-mediated functional analysis was conducted to assess the effects of changes in microbiota composition on microbiota gene function.Results indicated that metabolism was the dominant category associated with different treatments.Increased amino acid metabolism was observed in the CIP-treated ayu (Figure 7A),consistent with previous observations that antibiotic treatment can increase the relative concentrations of biogenic amines (derived from microbial amino acid metabolism) in the large intestine of antibiotictreated piglets (Mu et al.,2017).Increased energy metabolism duringP.plecoglossicidainfection,which is associated with reactive oxygen species (ROS) generation (Lin et al.,2020),may promote an immune response (Figure 7B).Compared to theP.pgroup,the co-administered group showed lower levels of energy and vitamin metabolism.Several studies have suggested that vitamin deficiency may lead to reduced resistance of the intestinal bacterial community to infection and increased risk to the host during pathogen invasion(Ribeiro et al.,2021).Hence,energy and vitamin metabolism deficiencies in the CIP-administered and infected group may attenuate the immune response toP.plecoglossicida.Consistently,compromised immune responses were observed in the primary immune tissues of infected ayu (Figure 9).
As crosstalk between intestinal microbiota,IECs,and immune responses during infection is essential for maintaining host homeostasis (Takiishi et al.,2017),changes in IECs and immune responses were evaluated in the different groups.CIP exposure induced severe IEC necrosis and reduced goblet cells (Figure 8A,CIP;8B,CIP+P.pand 8C),indicating that CIP accumulation may compromise the integrity and composition of the intestinal epithelial barrier.The intestines of theP.plecoglossicida-infected ayu were characterized by decreased villus height,increased villus width,and increased goblet cell number (Figure 8B,P.p).The intestines of the CIP andP.plecoglossicidaco-administered group showed complete destruction of the mucosal layer,severe epithelial cell necrosis,and reduction of goblet cells (Figure 8B,CIP+P.pand 8C).IECs mainly include ciliated,goblet,and basal cells.Goblet cells play a protective role by secreting mucin and anti-pathogenic compounds (e.g.,antimicrobial peptides,chemokines,and cytokines) into mucus,and forming goblet cell-associated antigen passages (GAPs) for adaptive immune responses (Peterson &Artis,2014).Based on immunohistochemical analysis,CIP exposure significantly decreased the number of goblet cells in the intestine,which may impair the anti-infection response ofP.plecoglossicidainfected ayu (Figure 8C).
As confirmed by the expression of pro-and antiinflammatory cytokines in the intestine (Figure 9A),gill(Figure 9B),spleen (Figure 9C),and head kidney (Figure 9D),CIP pre-exposure (CIP and CIP+P.pgroups) significantly down-regulated pro-inflammatory cytokines (IL-1βandTNF-α)and up-regulated anti-inflammatory cytokines (TGF-βandIL-10) in most tissues examined.In addition to its role as an antiinflammatory cytokine,TGF-βis a key regulator of the immune cells,epithelium,and microbiota in the intestine.Co-evolution of the intestinal microbiota and the host intestine has led to multiple mechanisms for the up-regulation of intestinalTGF-β.Epithelial cell injury can induceTGF-βproduction by IECs(Atarashi et al.,2013),andTGF-βlevels are elevated in patients with inflammatory bowel disease (McCabe et al.,1993),consistent with our observations that the CIP-treated groups showed more severe intestinal barrier damage (Figure 8A,B) and significantly up-regulatedTGF-βlevels (Figure 9A).The microbiota also regulates the production of intestinalTGFβ.For example,certain short-chain fatty acids produced byClostridiumspecies induce the production ofTGF-βby IECs(Martin-Gallausiaux et al.,2018).Here,Clostridium_sp._ID11(zOTU705) was significantly up-regulated in the CIP+P.pgroup (Figure 6B),further supporting the up-regulation of intestinalTGF-βduring CIP exposure.
The intestinal commensal microbiota is essential for appropriate systemic immune responses during pathogen infection (Ubeda &Pamer,2012).Microbiota-derived metabolites may have long-term effects on immune responses in systemic tissues.Short-chain fatty acids produced by the intestinal microbiota can enter tissues through blood circulation and inhibit neutrophil activation (Sun et al.,2017).Furthermore,microbial-derived ligands (such as lipopolysaccharide,CpG,and peptidoglycan) can activate immune responses in tissue-resident immune cells (Belkaid &Hand,2014).In our study,CIP exposure not only disrupted intestinal microbiota composition and intestinal barrier structure,but also impacted the immune responses of several immune tissues duringP.plecoglossicidainfection.CIP exposure significantly inhibitedIL-1β,TNF-α,andIL-10expression and promotedTGF-βexpression in most immune tissues (including the gill,spleen,and head kidney)(Figure 9B–D,CIP group),except for the increase inTNF-αand decrease inTGF-βfound in the spleen.When coadministered withP.plecoglossicida(CIP+P.pgroup),similar expression profiles were observed,except thatTGF-βwas down-regulated andIL-10was up-regulated in the head kidney.These results suggest that CIP pretreatment has profound effects on the immune response of systemic inflammatory tissues,which are only marginally affected by subsequent pathogen infection.This may be due to the negative regulatory roles of CIP-altered microbiota metabolites on systemic tissue immune responses,thus suppressing antipathogenic inflammatory responses.Further investigations are required to determine the mechanisms related to how CIP induces changes in microbiota,and which metabolic products contribute to intestinal barrier destruction and compromised systemic immune responses.
In addition to the effects of aquatic CIP overuse and accumulation on fisheries themselves,our study sheds some light on the effects of environmental CIP accumulation on the sustainable development of aquaculture and health of higher vertebrates,including humans.Environmental antibiotic residues have attracted worldwide attention.Notably,increasing concentrations of CIP have been detected in various aquatic matrices,including industrial wastewater effluent,hospital wastewater effluent,inland waters,marine systems,and groundwater (Booth et al.,2020;Kelly &Brooks,2018),reaching 245 550 ng/L in certain inland waters and 14 000 ng/L in groundwater in India (Fick et al.,2009;Kelly &Brooks,2018).With the extensive use of antibiotics in disease treatment,especially in agriculture and aquaculture,antibiotic consumption is expected to increase by 200% in 2030 (Klein et al.,2018),with water contamination at twice the current level due to the poor biodegradability of many antibiotics.We used a concentration of 1 mg/L CIP in the current study because it is near the high end of environmental levels and is likely to be close to the environmental level for the foreseeable future.Furthermore,previous studies have indicated that the effects of antibiotics on the fish gut microbiota are independent of the concentration of antibiotic used.Studies on zebrafish (Pindling et al.,2018),common carp (Zhao et al.,2021),Nile tilapia (Limbu et al.,2018),and large yellow croaker (Wang et al.,2020) have shown that higher concentrations of antibiotics exhibit the same trends in intestinal morphology,enzymatic activity,and RNA transcriptomes as observed in environmental concentration groups.Hence,our observations may represent what is happening in environmental water matrices now or in the near future in the absence of antibiotic use policies.The accumulated antibiotics in fish could also be transferred to terrestrial organisms,including humans,through predator-prey relationships,which may also increase the risks of pathogen infection.
CONCLUSIONS
This study is the first to report on the effects of CIP on intestinal homeostasis and systemic inflammatory immune response in subsequently infected aquatic organisms.Highdose CIP,as commonly given in aquaculture practice,significantly increased the mortality rate ofP.plecoglossicidainfected ayu by disrupting intestinal microbiota composition and intestinal barrier integrity and by compromising systemic immune responses (Figure 10).This research should broaden our understanding of the effects of antibiotic overuse on physiological activities of organisms,especially those under stress,and should enrich our knowledge of the side-effects of antibiotic overuse and discharge into waterways.
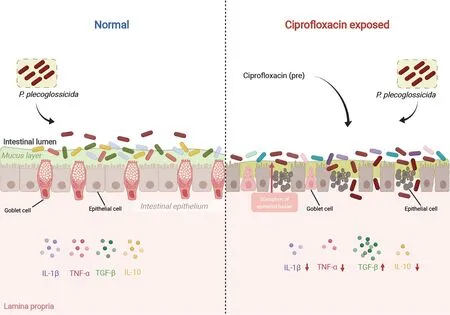
Figure 10 Effects of CIP contamination on intestinal homeostasis and immune responses of P.plecoglossicida-infected ayu
DATA AVAILABILITY
All sequences reported in this study were deposited in the Genome Sequence Archive database (http://gsa.big.ac.cn/)under accession No.CRA007443,Science Data Bank(https://www.scidb.cn/) under DOI:10.57760/sciencedb.01 947,and NCBI database under BioProjectID PRJNA825414.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
X.Y.W.conceptualization,methodology,software,and data curation.J.B.X.conceptualization,methodology,and software.C.J.F.writing original draft.T.D.,T.F.Z.,Z.Y.Z.,and J.P.investigation and resources.L.N.data curation,visualization,and writing original draft.J.C.supervision and writing &editing.All authors read and approved the final version of the manuscript.
- Zoological Research的其它文章
- Zoological Research call for papers of Cavefish Special Issue
- Fuel source shift or cost reduction:Context-dependent adaptation strategies in closely related Neodon fuscus and Lasiopodomys brandtii against hypoxia
- Ecological study of cave nectar bats reveals low risk of direct transmission of bat viruses to humans
- Population and conservation status of a transboundary group of black snub-nosed monkeys (Rhinopithecus strykeri) between China and Myanmar
- Nucleus accumbens-linked executive control networks mediating reversal learning in tree shrew brain
- Europe vs.China:Pholcus (Araneae,Pholcidae) from Yanshan-Taihang Mountains confirms uneven distribution of spiders in Eurasia

