Rice MtN3/saliva/SWEET gene family:Evolution,expression profiling,and sugar transport
Meng Yuan,Junwei Zhao,Renyan Huang,Xianghua Li,Jinghua Xiao and Shiping Wang
National Key Laboratory of Crop Genetic Improvement,National Center of Plant Gene Research(Wuhan),Huazhong Agricultural University,Wuhan 430070,China.*Correspondence:swang@mail.hzau.edu.cn
INTRODUCTION
MtN3/saliva/SWEET-type genes,encoding transmembrane proteins,are prevalent in different types of cellular organisms(Yuan and Wang 2013).The first MtN3/saliva/SWEET-type gene,MtN3,was identified in the legume Medicago truncatula as being associated with different stages of Rhizobium-induced nodule development(Gamas et al.1996).Later,another MtN3/saliva/SWEET-type gene,saliva,was identified in Drosophila,which had a high transcript level in the salivary gland during all subsequent stages of embryonic development(Artero et al.1998).Recently,this type of protein was also named SWEET because some of the proteins perform as sugar transporters(Chen et al.2010).These proteins feature the MtN3/saliva(MtN3_slv)domain,which was later renamed as the PQ loop repeat.It comprises a pair of repeats,each spanning two transmembrane helices connected by a loop based on the description of the Conserved Domain Database(http://www.ncbi.nlm.nih.gov/cdd).However,it has been shown that one of the two MtN3/saliva domains in rice Xa13/Os8N3/SWEET11 consists of three transmembrane helices connected by two loops(Yuan et al.2010).Most MtN3/saliva/SWEET proteins are predicted to harbor two MtN3/saliva domains,and a few harbor one MtN3/saliva domain(Yuan and Wang 2013).
In contrast to mammals,which have only one MtN3/saliva/SWEET-type gene,plants carry multiple MtN3/saliva/SWEET-type genes in each genome,suggesting their importance in plants(Yuan and Wang 2013).Some plant MtN3/saliva/SWEET-type genes have been reported to be involved in different biological activities;for example,host–pathogen interactions,reproductive development,senescence,and abiotic stress responses(Yuan and Wang 2013).
The rice MtN3/saliva/SWEET gene family consists of 21 paralogs(Yuan and Wang 2013).The physiological functions of only two paralogs,Xa13/Os8N3/SWEET11 and Os11N3/SWEET14,are known,although expression profiling of a number of rice MtN3/saliva/SWEET-type genes has been reported and suggests putative roles in various biological activities(Yuan and Wang 2013).Xa13/Os8N3/SWEET11-suppressed plants show reduced fertility or are sterile due to compromised microspore development,which results in the gradual degeneration of immature pollens.This suggests that Xa13/Os8N3/SWEET11 is indispensable for rice pollen development(Chu et al.2006).Os11N3/SWEET14-knockout mutants show reduced seed size and delayed reproductive development(Antony et al.2010).
Interestingly,the pathogenic bacterium Xanthomonas oryzae pv.oryzae(Xoo),which causes bacterial blight,one of the most devastating rice diseases worldwide,seems to like using MtN3/saliva/SWEET-type genes to facilitate its invasion of rice plants.Different Xoo strains specifically activate the expression of Xa13/Os8N3/SWEET11,Os11N3/OsSWEET14,or another rice MtN3/saliva/SWEET-type gene,Xa25/SWEET13,to facilitate infection(Chu et al.2006;Yang et al.2006;Antony et al.2010;Liu et al.2011).Mimicking Xoo-induced expression of rice SWEET12 also increases rice susceptibility to this bacterium(Li et al.2013).It has been demonstrated that Xoo strain PXO99,which transcriptionally induces Xa13/Os8N3/SWEET11,is sensitive to copper(Yuan et al.2010).Xa13/Os8N3/SWEET11 cooperates with two copper transporter-type proteins,COPT1 and COPT5,to remove copper from xylem vessels where Xoo multiplies and spreads to cause disease.Induction of Xa13/Os8N3/SWEET11 by PXO99 results in reduced copper content in xylem vessels,which facilitates the invasion of this bacterium(Yuan et al.2010).Xa13/Os8N3/SWEET11 was able to function as a low-affinity glucose and sucrose transporter in a human cell line and Xenopus oocytes,and the same activity was shown for Os11N3/SWEET14;these results suggested that the two proteins move sugar from cells to the apoplasm to provide nutrients to Xoo for its growth and virulence(Chen et al.2010).
The Oryza genus consists of two cultivated rice species,Asian cultivated rice O.sativa and African cultivated rice O.glaberrima,both of which carry the AA genome,and 22 wild rice species,representing AA,BB,CC,BBCC,CCDD,EE,FF,GG,HHJJ,and KKLL genomes(Ge et al.1996;Ammiraju et al.2006).To further understand the biological roles of rice MtN3/saliva/SWEET family paralogs,we analyzed their evolution by examining their presence in wild rice species;their expression profiles in selected tissues during different developmental stages and under varied hormone-dependent signaling;and their putative functions in sugar transport.The results suggested that rice MtN3/saliva/SWEET-type genes have diversified expression patterns and may be involved in multiple functions in plant growth,development,and response to environmental stresses.
RESULTS
Approximately half of rice MtN3/saliva/SWEET paralogs predate the differentiation of Oryza genomes
Based on bioinformatic analysis,18 of the 21 rice MtN3/saliva/SWEET paralogs from indica rice(O.sativa ssp.indica)varieties IR24(Xa13/Os8N3/SWEET11)and Zhenshan 97(Xa25/SWEET13)and japonica rice(O.sativa ssp.japonica)variety Nipponbare(rest of the 18 paralogs)encode proteins with two MtN3/saliva domains;Os01g40960,Os09g08030/SWEET7a,and Os09g08270/SWEET7e from Nipponbare each harbor only one MtN3/saliva domain(Figures 1A,S1;Table S1).The 21 paralogs are distributed on eight of the 12 rice chromosomes(Figure 1B).Different phylogenetic analysis programs(neighbor-joining,maximum likelihood,and maximum parsimony)were carried out to examine the evolutionary relationship within the rice MtN3/saliva/SWEET family.Similar results were obtained from these analyses when using full-length sequences of these proteins as matrix(Treebase accession no.S15061;http://purl.org/phylo/treebase/phylows/study/TB2:S15061).Seven pairs of paralogs,Os09g08440/SWEET7b–Os09g08270/SWEET7e,Os09g08030/SWEET7a–Os12g07860/SWEET7c,Os01g42110/SWEET6a–Os01g42090/SWEET6b,Os05g12320/SWEET3a–Os01g12130/SWEET3b,Os01g36070/SWEET2a–Os01g50460/SWEET2b,Os01g65880/SWEET1a–Os05g35140/SWEET1b,and Xa25/SWEET13–Os11N3/SWEET14,were always classified into the same clades with high bootstrap values(≥96 in at least two analysis methods)no matter whether using neighborjoining,maximum likelihood or maximum parsimony method(Figures 1C,S2).When using the Gblocks method to eliminate poorly aligned and highly variable positions of the MtN3/saliva/SWEET protein sequences(Talavera and Castresana 2007),the largest aligned segments were at the amino-terminal regions of the 21 proteins,which consisted of approximately 50 amino acids for most of the aligned sequences(Figure S3;Treebase accession no.S15061).The phylogenetic trees constructed using the highly conserved amino-terminal regions with the three analysis methods show average lower bootstrap values compared with the trees constructed using the full-length sequences(Figure S4).However,except for the Os09g08440/SWEET7b–Os09g08270/SWEET7e pair,the other six pairs of paralogs listed above were also classified into the same clades with bootstrap values 76 or more in at least two analysis methods.The similar chromosomal locations and close phylogenetic relationship suggest that the Xa25/SWEET13–Os11N3/SWEET14 pair,the Os01g12130/SWEET3b–Os05g12320/SWEET3a pair,the Os01g65880/SWEET1a–Os05g35140/SWEET1b pair,and the Os09g08030/SWEET7a–Os12g07860/SWEET7c pair could be generated by chromosomal fragmental duplication during evolution(Figures 1B,C,S2,S4).In addition,the tandem locations and close phylogenetic relationship suggest that the Os01g42090/SWEET6b and Os01g42110/SWEET6a pair could be generated by direct tandem duplication during evolution(Figures 1B,C,S2,S4).These inferences are also supported by the similar protein and gene structures of each of these pairs(Figures 1A,S1).
To further analyze the evolution of the rice MtN3/saliva/SWEET gene family,we examined the distribution of this type of gene in 22 wild rice accessions(Table S2),which included seven accessions of O.rufipogon(AA genome),two accessions of O.punctata(BB),two accessions of O.officinalis(CC),one accession of O.minuta(BBCC),three accessions of O.alta(CCDD),two accessions of O.grandiglumis(CCDD),three accessions of O.latifolia (CCDD),one accession of O.australiensis(EE),and one accession of O.meyeriana(GG),by polymerase chain reaction(PCR)amplification analysis using three pairs of primers,the amplified products of which partially overlapped for each paralog(Table S3).Partial genomic sequences of all of the analyzed wild rice species were available in the High-Throughput Genomic Sequences(HTGS) database (http://www.ncbi.nlm.nih.gov/genbank/htgs).The HTGS database was searched using the DNA sequences of all of the rice MtN3/saliva/SWEET paralogs as queries by BLAST analysis(Altschul et al.1997).Each MtN3/saliva/SWEET paralog existed in wild rice accessions could be amplified by using all of the three pairs of primers designed,but only one amplification fragment was presented(Figure 2).The paralog is considered not present in the wild rice accession,if its homolog cannot be amplified by using any of the three pairs of primers from corresponding DNA template and its homolog is not detected in the genomic sequences of corresponding wild rice species by BLAST analysis.
The homologs of eight rice MtN3/saliva/SWEET paralogs(Os01g36070/SWEET2a, Os02g30910/SWEET15, Os03g22590/SWEET12,Xa13/Os8N3/SWEET11,Os09g08270/SWEET7e,Os11N3/SWEET14,Os12g07860/SWEET7c,and Xa25/SWEET13)were detected in all 22 wild rice accessions from all eight species examined(Figure 2;Table S2).The homologs of the other two rice MtN3/saliva/SWEET paralogs(Os01g65880/SWEET1a and Os05g12320/SWEET3a)were detected in all eight wild rice species,although not all of the accessions examined.The homolog of Os03g22200/SWEET16 was detected in all of the examined wild rice genome types except GG,and the homolog of Os05g51090/SWEET5 was detected in all of the examined wild rice genome types except EE and GG.The homologs of the remaining nine rice MtN3/saliva/SWEET paralogs were detected in only one to four wild rice genome types(AA,BB,CC,or BBCC;Figure 2).The GG genome occupies the most basal position of the Oryza genus,and it is considered to be the most distantly related to cultivated rice(Ge et al.1996).These results suggest that approximately half of the paralogs in rice MtN3/saliva/SWEET family may originate before the differentiation of other genome types in the Oryza genus.

Figure 1.Analysis of the evolution of rice MtN3/saliva/SWEET family(A)The predicted structures of rice MtN3/saliva/SWEET proteins.(B)The distribution of MtN3/saliva/SWEET paralogs on rice chromosomes.The centromere of each chromosome is indicated with a black circle.(C)Phylogenetic relationship of rice MtN3/saliva/SWEET proteins analyzed by neighbor-joining method.The numbers for interior branches indicate the bootstrap values(%)for 1,000 replications.The scale bar represents 0.2 amino acid substitution per site.

Figure 2.Distribution of MtN3/saliva/SWEET-type genes in wild rice species analyze by polymerase chain reaction(PCR)amplification analysisThe wild rice species represented by the material number are listed in Table S2.AA,BB,CC,BBCC,EE,and GG are different rice genomes.
MtN3/saliva/SWEET paralogs differentially expressed in vegetative and reproductive tissues
To study the potential biological roles of different rice MtN3/saliva/SWEET paralogs,we analyzed their transcription profiles in root,stem,flag leaf,sheath,flower,and panicle branch tissues at the booting(panicle development)stage(Figure 3).Among the 21 paralogs,15 were expressed in all six tissues examined,and the other six were expressed in one to four of the six tissues.Os03g22590/SWEET12 and Os09g08030/SWEET7a showed the most restricted expression,and their transcripts were only detected in roots.The transcripts of 17 paralogs were detected in flowers,although with different expression levels.These results suggest that the MtN3/saliva/SWEET gene family may play important roles in rice reproductive development in addition to having a function in non-reproductive tissues.
Most of the reported MtN3/saliva/SWEET-type genes from plants and lower eukaryotes are associated or putatively associated with reproductive development(Yuan and Wang 2013).To gain further insight into the potential biological roles of MtN3/saliva/SWEET genes in reproductive development,we analyzed the expression of the 17 paralogs in flower development processes that were represented by panicle length(~0.5–22 cm).The 17 paralogs showed four types of expression patterns in flower development(Figure 4).The paralogs of pattern I showed a relatively consistent low level of expression in the developing flowers during the growth of panicles from approximately 0.5–22 cm.The expression levels of pattern II paralogs had a gradual increase followed by a gradual reduction to the basal level during development of the panicle.The transcripts of pattern III paralogs increased gradually during panicle development and reached the highest level at approximately the 22 cm panicle stage.The expression levels of pattern IV paralogs were relatively consistent in panicle development from approximately 0.5–11 cm and decreased afterward.These results suggest that different paralogs may be associated with discrete stages of reproductive development.
Leaf senescence is the final stage of leaf development.The expression of an Arabidopsis MtN3/saliva/SWEET-type gene(SAG29/SWEET15)increased gradually during leaf senescence,and transgenic plants overexpressing SAG29/AtSWEET15 showed accelerated senescence(Quirino et al.1999;Seo et al.2011).To examine whether rice MtN3/saliva/SWEET genes are also associated with leaf senescence,we comparatively analyzed their expression in young and naturally senesced leaves.It is known that natural leaf senescence is accompanied by reduced chlorophyll content(Lim et al.2007).In addition,rice OsDOS and Osl57 are marker genes of leaf senescence.OsDOS negatively regulates leaf senescence(Kong et al.2006),while Osl57 expression is induced during leaf senescence,suggesting that it may be coordinately regulated by a senescence-inducing factor during rice leaf senescence(Lee et al.2001).In this study,we used chlorophyll content and the expression levels of OsDOS and Osl57 to evaluate the status of leaf senescence.Young leaves,moderately senesced leaves,and senesced leaves were used for gene expression analysis(Figure 5A).The reduction of chlorophyll content was associated with the level of leaf senescence based on comparison with the chlorophyll content in young leaves(Figure 5B).OsDOS and Osl57 showed gradually suppressed and induced expression as leaf senescence progressed,respectively(Figure 5C).The expression of only 18 of the 21 MtN3/saliva/SWEET paralogs,except for Os01g42110/SWEET6a,Os09g08030/SWEET7a,and Os09g08440/SWEET7b,was detected in flag leaves in the present experimental condition(Figure 5D).Six of them showed a similar expression pattern as OsDOS in different developed rice leaves,whereas another five paralogs showed a similar expression pattern as Osl57 in different developed leaves(Figure 5D).The remaining seven paralogs presented no obvious differences in expression in rice leaves at different developmental stages.These results suggest that some of the rice MtN3/saliva/SWEET paralogs may be negatively or positively involved in the regulation of leaf senescence.

Figure 3.Tissue-specific expression patterns of rice MtN3/saliva/SWEET paralogs analyzed by reverse transcription polymerase chain reaction(RT-PCR)Samples were collected from japonica rice varieties Zhonghua 11 at the booting(panicle development)stage in which the panicle was visible as a white feathery cone to the naked eye.The Arabic numerals on the right side are PCR amplification cycles.
A set of MtN3/saliva/SWEET paralogs transcriptionally responded to phytohormones
Phytohormones play critical roles in various biological activities.To investigate potential involvement of MtN3/saliva/SWEET paralogs in phytohormone signaling,we performed a comprehensive expression analysis of these paralogs in rice by processing genome-wide microarray data collected after phytohormone treatment.Affymetrix genechip rice genome array probes for 18 of the 21 MtN3/saliva/SWEET paralogs were identified in the microarray database(http://www.ncbi.nlm.nih.gov/;accession no.GSE19024)(Wang et al.2010);the genechip array did not include the probes for Os01g42110/SWEET6a,Os09g08030/SWEET7a,and Os09g08440/SWEET7b(Table S1).After treatment of rice seedlings with gibberellic acid 3(GA3),cytokinin,or 1-naphthalene acetic acid(NAA;an analog of indole-3-acetic acid,the major type of auxin in rice),a set of MtN3/saliva/SWEET paralogs showed differential expression compared to the untreated control(Figure S5).The expression of five paralogs(Os01g12130/SWEET3b,Os02g30910/SWEET15,Os05g12320/SWEET3a,Os01g50460/SWEET2b,and Os05g35140/SWEET1b)was obviously induced after GA3,cytokinin,or NAA treatment,whereas the expression of Os01g65880/SWEET1a and Os02g19820/SWEET4 was slightly suppressed after GA3,cytokinin,or NAA treatment.In addition,Os09g08270/SWEET7e was induced after either GA3 or cytokinin treatment,Os05g51090/SWEET5 was induced after NAA treatment,Os11N3/SWEET14 was slightly induced after either GA3 or NAA treatment,and Os03g22590/SWEET12 was suppressed after either GA3 or NAA treatment(Figure S5).These results suggest that some MtN3/saliva/SWEET paralogs may be involved in biological activities regulated by GA,cytokinin,or auxin.
A set of MtN3/saliva/SWEET paralogs functioned as sugar transporters in yeast
Recent studies have reported that the Xa13/Os8N3/SWEET11 and Os11N3/SWEET14 function as low-affinity glucose and sucrose transporters in mammalian cells and oocytes(Chen et al.2010,2012).To investigate the potential roles of the other rice MtN3/saliva/SWEET paralogs in sugar(monosaccharide or disaccharide)transport,the selected 11 paralogs were expressed separately in yeast(Saccharomyces cerevisiae)mutant EBY.VW4000,which lacked sugar uptake ability,under the control of the promoter of S.cerevisiae high-affinity hexose transporter HXT7 gene(Wieczorke et al.1999;Ye et al.2001).All transformants grew on synthetic-deficient(SD)media containing 2%maltose,indicating the presence of the expression vector or target gene(Figure 6).The lack of growth of EBY.VW4000 cells containing empty vector(negative control)was restored when transformed with yeast HXT7(positive control)in the medium supplemented with 2%glucose,fructose,mannose,galactose,or sucrose.The transformants carrying any one of the 11 rice paralogs did not grow in the medium supplemented with glucose,fructose,mannose,or sucrose,indicating that the proteins encoded by the 11 paralogs cannot transport these four sugarsinyeast.However,expressionofriceOs02g19820/SWEET4,Os02g30910/SWEET15,Os05g35140/SWEET1b,or Os05g51090/SWEET5 alone could efficiently complement the phenotype of EBY.VW4000 mutant in the medium supplemented with galactose as compared to the positive control,suggesting that these four rice proteins function as high-affinity galactose transporters in yeast.

Figure 4.Expression patterns of MtN3/saliva/SWEET paralogs in developing flowers analyzed by reverse transcription polymerase chain reaction(RT-PCR)Samples were collected from different sized panicles of japonica rice varieties Zhonghua 11 at the booting stage.Panicle(P)size P1,~0.5 cm;P2,~1.0 cm;P3,~2.0 cm;P4,~3.5 cm;P5,~4.5 cm;P6,~11 cm;P7,~16.5 cm;P8,~19 cm;P9,~22 cm.According to the relationship between panicle length and stage of panicle development,0–3 cm panicle is at floral transition to floral organ development stages,3–10 cm panicle is at meiotic stage,10–15 cm panicle is at young microspore stage,and 15–22 cm panicle is at vacuolated pollen stage.The Arabic numerals on the right side are PCR amplification cycles.
DISCUSSION
Although MtN3/saliva/SWEET-type genes are prevalent in cellular organisms,the roles in physiological processes and the biochemical functions of their encoded proteins are largely unknown in rice(Yuan and Wang 2013).The present results provide further insight into the potential roles in development and the putative biochemical function as sugar transporters of some rice MtN3/saliva/SWEET paralogs.
One third of rice MtN3/saliva/SWEET paralogs may originate recently
The MtN3/saliva/SWEET-type genes are evolutionarily conserved.However,the number of genes of this type within a species is not always associated with the evolutionary complexity of the species.For example,some bacteria,chordates,and mammals have only one MtN3/saliva/SWEET-type gene,but an arthropod(Drosophila melanogaster)has two MtN3/saliva/SWEET-type genes,and an Aschelminth(Caenorhabditis elegans)has seven MtN3/saliva/SWEET-type genes(Yuan and Wang 2013).The present results suggest that the number of rice MtN3/saliva/SWEET paralogs gradually increased with the evolution of the rice genome.

Figure 5.Analysis by quantitative reverse transcription polymerase chain reaction(qRT-PCR)revealed that expression of some MtN3/saliva/SWEET paralogs was influenced during natural leaf senescenceRice variety Zhonghua 11 were grown in a greenhouse.Samples were collected from 10 week old plants.YL,young leaf(black histogram);MSL,moderately senesced leaf(black stripe histogram);SL,senesced leaf(black dot histogram).Asterisks indicate that a significant difference was detected between YL and MSL or SL at**P<0.01 and*P<0.05.(A)The appearance of rice leaves at different developmental stages.(B)The chlorophyll content in rice leaves at different developmental stages.(C)Expression levels of senescence-related genes,OsDOS and Osl57,in rice leaves at different developmental stages.(D)Expression patterns of MtN3/saliva/SWEET paralogs in rice leaves at different developmental stages.
The phylogeny of Oryza genomes shows that the AA genome is most closely related to the BB genome and the relationship gradually declines in the following order:BBCC,CC,CCDD,EE,KK,HHKK,FF,HH,HHJJ,JJ,and GG genomes.The GG genome,which diversified approximately 13–14.6 million years ago,occupies the most basal position of the genus,and it is the most distantly related to the AA genome(Ge et al.1996;Ammiraju et al.2008).The homologs of 10 rice MtN3/saliva/SWEET paralogs(Os01g36070/SWEET2a,Os02g30910/SWEET15,Os03g22590/SWEET12,Xa13/Os8N3/SWEET11,Os09g08270/SWEET7e,Os11N3/SWEET14,Os12g07860/SWEET7c,Xa25/SWEET13,Os01g65880/SWEET1a,and Os05g12320/SWEET3a)were detected in the GG genome in addition to the other wild rice genomes examined(Figure 2).Analysis of the phylogenetic relationship between rice and Arabidopsis MtN3/saliva/SWEET families have revealed that the 10 rice MtN3/saliva/SWEET paralogs are classified into three clades with each clade containing Arabidopsis MtN3/saliva/SWEET paralogs(Chen et al.2010).These results suggest that the 10 rice paralogs may have originated before the speciationofmonocots and dicots.
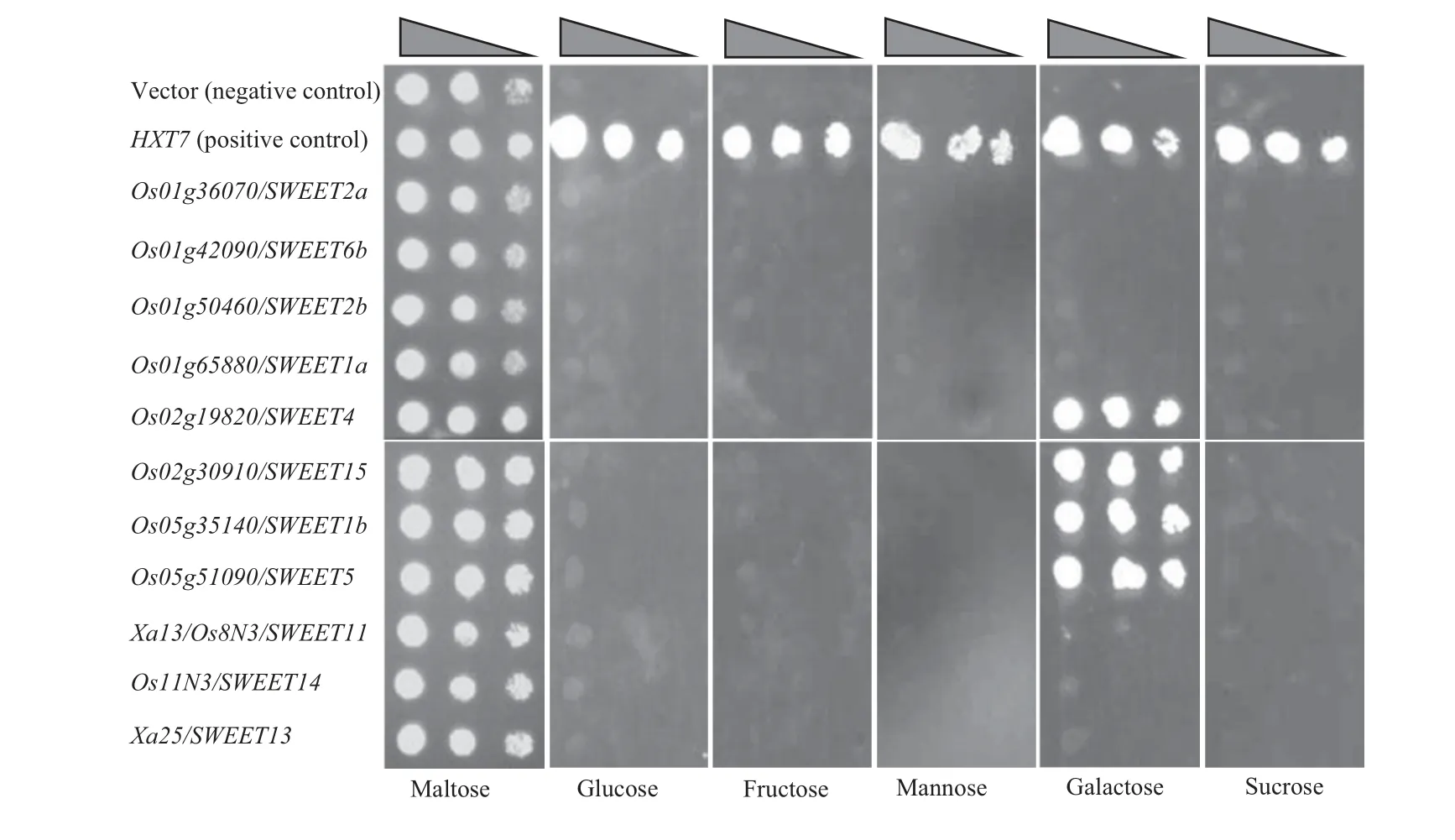
Figure 6.Functional complementation of Saccharomyces cerevisiae EBY.VW4000 mutant by expression of rice MtN3/saliva/SWEET paralogsComplementation is indicated by cell growth on the media supplemented with 2%glucose,fructose,mannose,galactose,or sucrose.Yeast HXT7 gene was used as a positive control.Empty vector was used as a negative control.Triangle indicates 10-fold diluted cells were spotted onto synthetic-deficient media plates.
The homologs of the remaining 11 rice MtN3/saliva/SWEET paralogs were only detected in wild rice with AA,BB,CC,BBCC,CCDD,or EE genomes but not the GG genome,indicating that these rice paralogs likely originated after the formation of the Oryza genus.The AA genome appeared approximately 0.58 million years ago(Ammiraju et al.2008).Thus,Os01g12130/SWEET3b,Os05g35140/SWEET1b,Os09g08440/SWEET7b,Os02g19820/SWEET4,Os01g40960,Os01g42090/SWEET6b,and Os01g42110/SWEET6a are young paralogs based on the distribution of their homologs in the 22 wild rice accessions examined(Figure 2).
Segmental duplications are a common event in the rice genome,which results in a homologous gene family in the same locations or stretches of different chromosomes(Wang et al.1999,2000).According to phylogenic and chromosomal location analyses,the Os09g08030/SWEET7a and Os12g07860/SWEET7c pair,the Os01g12130/SWEET3b and Os05g121320/SWEET3a pair,and the Os01g65880/SWEET1a and Os05g35140/SWEET1b pair may be generated by chromosomal fragmental duplications followed by divergence of the duplicated genes during evolution(Figure 1B,C).If this prediction is correct,it is clear that Os09g08030/SWEET7a may originate from ancient Os12g07860/SWEET7c predating the differentiation of BB and CC genomes,which occurred approximately 5.7–7.5 million years ago,and Os01g12130/SWEET3b and Os05g35140/SWEET1b may originate from ancient Os05g121320/SWEET3a and Os01g65880/SWEET1a,respectively,predating the origin of cultivated rice,which happened approximately 10 000–15 000 years ago(Khush 1997;Ammiraju et al.2008).Furthermore,the tandem location and close phylogenetic relationship of Os01g42090/SWEET6b and Os01g42110/SWEET6a indicates that this pair of paralogs may have been recently generated by direct tandem duplication(Figure 1B,C).However,the predicted duplicate gene pairs did not show the similar tissue expression patterns each other(Figures 3,4;Yuan and Wang 2013).Thus,further study is required to examine the prediction of MtN3/saliva/SWEET family evolution and the putative diversification of promoters between duplicate genes.
Rice MtN3/saliva/SWEET paralogs may be associated with multiple physiological processes
So far,most of the reported MtN3/saliva/SWEET-type genes from different species have been associated or putatively associated with reproductive development(Yuan and Wang 2013).The present results provide further evidence to support the observation.In addition to Xa13/Os8N3/SWEET11 and Os11N3/SWEET14 being required for normal rice reproductive development(Chu et al.2006;Antony et al.2010),the present results show that most(17)of the rice MtN3/saliva/SWEET paralogs are expressed in flower tissues and approximately half of the paralogs(expression patterns II–IV),including Xa13/Os8N3/SWEET11 and Os11N3/SWEET14,are differentially expressed during panicle development(Figure 4).Of the 17 paralogs,10 were evolutionarily conserved and their homologs occurred in all of the wild rice genome types,although not all of the 22 wild rice accessions examined in this study.These results suggest that rice MtN3/saliva/SWEET family may be tightly associated with reproductive development,and different paralogs of this family may be specifically involved in different developmental stages.Interestingly,the ancestors of three young paralogs(Os02g19820/SWEET4,Os05g35140/SWEET1b,and Os01g40960),which were generated only after the appearance of the AA genome,also showed differential expression in flowers during panicle development(Figures 2,4).The AA genome,which includes cultivated rice,has the widest geographic distribution and is well adapted compared to any wild rice genomes throughout the world(Ge et al.1996).Thus,it is worth elucidating whether the origin of the three paralogs is associated with these features.
Only one MtN3/sliva/SWEET-type gene,Arabidopsis SAG29/AtSWEET15,has been reported to be associated with senescence so far(Quirino et al.1999;Seo et al.2011).However,the present results show that 11 rice paralogs are differentially expressed in senesced leaves compared to their expression in young leaves(Figure 5).Six(Os01g65880/SWEET1a, Os01g36070/SWEET2a, Os01g12130/SWEET3b,Os01g42090/SWEET6b,Os02g19820/SWEET4,and Os05g35140/SWEET1b)of the 11 paralogs showed a similar expression pattern as OsDOS,the marker gene of a negative regulator for leaf senescence(Kong et al.2006),suggesting that the six paralogs may be negatively associated with rice leaf senescence.Interestingly,Os01g65880/SWEET1a and Os05g35140/SWEET1b are predicted as duplicate genes as discussed in the previous section,suggesting that some duplicate genes may have conserved physiological functions,although they show diversification in tissue expression patterns.Another five paralogs(Os01g40960,Os01g50460/SWEET2b,Os02g30910/SWEET15,Os05g51090/SWEET5,and Xa13/Os8N3/SWEET11)showed a similar expression pattern as Osl57,the marker gene of a putative positive regulator for leaf senescence(Lee et al.2001),suggesting that the five paralogs may be positively associated with rice leaf senescence.However,unlike the reproductive process,in which most of the putatively involved MtN3/saliva/SWEET paralogs appear to predate the formation of the Oryza genus,seven of the 11 paralogs originated after the differentiation of EE and GG genomes.Among the seven,five(Os01g12130/SWEET3b,Os05g35140/SWEET1b,Os02g19820/SWEET4,Os01g40960,and Os01g42090/SWEET6b)originated after the differentiation of AA and BB genomes,which was approximately 2 million years ago(Kim et al.2007).These results suggest that regulation of senescence may gradually become more important during evolution.
The tissue-specific expression patterns(Figure 3)suggest that the functions of some rice MtN3/saliva/SWEET paralogs may be associated with root or stem tissues.In addition,some paralogs also may be involved in the regulation of physiological processes depending on the signaling of GA,cytokinin,or auxin,which regulate plant growth and developmental processes as well as biotic and abiotic stress responses(Bari and Jones 2009).It is obvious that some MtN3/saliva/SWEET paralogs may be associated with multiple physiological processes,such as both reproductive development and leaf senescence.The expression specificity of the MtN3/saliva/SWEET family will provide guidance for further characterization of each paralog in rice development,growth,environmental adaptation,rice-pathogen interaction,and so forth.
Rice MtN3/saliva/SWEET paralogs may have different biochemical functions
MtN3/saliva/SWEET-type proteins have been suggested to be sugar transporters,and thus they are named SWEET(Chen et al.2010,2012).The present results show that four rice MtN3/saliva/SWEET paralogs (Os02g19820/SWEET4,Os02g30910/SWEET15,Os05g35140/SWEET1b,and Os05g51090/SWEET5)can function as galactose-specific high-affinity transporters in yeast cells(Figure 6).However,Xa13/Os8N3/SWEET11 and Os11N3/SWEET14,which function as low-affinity glucose and sucrose transporters in animal cell line and oocytes(Chen et al.2010,2012),could not transport glucose,sucrose,or other sugars in yeast in the present experimental condition.This divergence may be due to the different experimental systems employed.Thus,further studies are required to characterize the sugar transporter features of rice MtN3/saliva/SWEET paralogs in plants.
The present results reveal that not every rice MtN3/saliva/SWEET paralog can function as a sugar transporter in yeast cells,which suggests that MtN3/saliva/SWEET-type proteins may have other biochemical functions.This inference is supported by evidence that Xa13/Os8N3/SWEET11 physically interacts with two copper transporter-type proteins COPT1 and COPT5,and the three proteins collectively remove copper from xylem vessels in the rice-Xoo interaction;this feature of Xa13/Os8N3/SWEET11 was used by the copper-sensitive Xoo strain to invade rice(Yuan et al.2010).Because Xa13/Os8N3/SWEET11 does not possess the structural features of a copper transporter(Nose et al.2006)and other rice COPT proteins can transport copper alone or in heterodimer form in yeast(Yuan et al.2011),it is speculated that this protein may function in help the trafficking of COPT1 and COPT5 to the plasma membrane or the stabilization of the COPT1–COPT5 complex on the plasma membrane for copper influx in rice(Yuan et al.2010;Yuan and Wang 2013).
MATERIALS AND METHODS
Wild rice materials
All of the wild rice seeds or DNA used in this study were from our collection(Xiao et al.2009)or provided by the International Rice Research Institute(Table S2).These materials included the species Oryza rufipogon(AA genome),O.punctata(BB genome),O.officinalis(CC genome),O.minuta(BBCC genome),O.alta(CCDD genome),O.grandiglumis(CCDD genome),O.latifolia(CCDD genome),O.australiensis(EE genome),and O.meyeriana(GG genome).Polymerase chain reaction primers used to analyze MtN3/saliva/SWEET-type genes in wild rice materials are listed in Table S3.
Sequence analysis
The MtN3/saliva domain of proteins was analyzed by searching the Conserved Domain Database(http://www.ncbi.nlm.nih.gov/cdd).Multiple sequence alignments of the amino acid sequences were generated using ClustalW in MEGA 5.1 with the default parameters(Tamura et al.2011).The obtained sequence alignments were used as the input for the neighbor joining or maximum parsimony in MEGA 5.1,or maximum likelihood in PhyML to construct the phylogenetic tree(Guindon et al.2010).For phylogenetic tree construction,bootstrap method and interior-branch test with 1,000 replications were used for test of phylogeny,with Poisson model and pairwise deletion during gaps/missing date treatment.
Gene expression analysis
Total RNA from different tissues of japonica rice(O.sativa ssp.japonica)variety Zhonghua 11 at different developmental stages was used for gene expression analyses by reverse transcription(RT)-PCR or quantitative(q)RT-PCR as described previously(Zhou et al.2002;Qiu et al.2007).Polymerase chain reaction primers for RT-PCR and qRT-PCR are listed in Table S4.The priming efficiency of each pair of primers used for qRT-PCR was 96%–104%by ABI PRISM 7500 Real-Time PCR System SDS Software version 1.4(Applied Biosystems,Foster City,CA,USA).The expression level of the rice actin gene was first used to standardize the RNA sample for each qRT-PCR.The expression level relative to control was then presented.Each qRT-PCR or RT-PCR assay was repeated at least twice with similar results,with each repetition having three replicates.
Chlorophyll assays
Leaves of rice plants at different developmental stages were used for chlorophyll measurements.The chlorophyll was extracted in 100%ethanol,and chlorophyll content was determined photometrically using the method of Wintermans and De Mots(1965).
Plasmid construction
The 708 bp promoter of S.cerevisiae HXT7 gene was amplified from plasmid p426-pHXT7-HXT7(Hamacher et al.2002)using specific primers(Table S5)and subcloned into the SacI/XbaI site of the p413GPD(+His)vector to replace the glyceraldehyde-3-phosphate dehydrogenase promoter as described previously(Mumberg et al.1995),which resulted in the plasmid p413HXT7.The S.cerevisiae HXT7 gene was amplified from the plasmid p426-pHXT7-HXT7 using gene-specific primers(Table S5)and was inserted into the BamHI/EcoRI sites of the p413HXT7 vector.The full-length cDNAs of rice MtN3/saliva/SWEET paralogs were obtained by RT-PCR using gene-specific primers(Table S5)from rice variety Zhonghua 11.The cDNAs of Os01g36070,Os01g42090,Os01g65880,Os02g19820,Os02g30910,Os05g35140,Xa13/Os8N3/SWEET14,Os11N3/SWEET11,and Xa25/SWEET13 were subcloned into the BamHI/EcoRI sites of p413HXT7,respectively.The cDNAs of Os01g50460 and Os05g51090 were subcloned into the SpeI/EcoRI sites of p413HXT7.All cDNA sequences were confirmed by DNA sequencing.
Functional complementation analyses in yeast
To study the putative function of rice MtN3/saliva/SWEET genes in monosaccharide or disaccharide transport,a plasmid carrying the target gene was transformed into the yeast strain EBY.VW4000(MATa Δhxt1-17 Δgal2 Δstl1 Δagt1 Δmph2 Δmph3 leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8cSUC2)(Wieczorke et al.1999)by the lithium acetate procedure(Ito et al.1983).Synthetic media consisted of 6.7 g/L Difco yeast nitrogen base(YNB;BD Biosciences,San Jose,CA,USA)supplemented with amino acids but without histidine as a plasmid-selection marker were used for plasmid transformation into yeast cells.For complementation assays,the transformed yeast cells were grown overnight in liquid minimum(SC-His)medium to exponential phase(OD600nm=1.0).Several 10-fold diluted yeast cells were plated as drops on SD media containing either 2%maltose(as control)or 2%glucose,2%fructose,2%mannose,2%galactose,or 2%sucrose,plus respective auxotrophic requirements.Plates were incubated for 3–5 d at 30 °C and documented by scanning.
Statistical analysis
The significance of differences between samples was analyzed by the pairwise Student’s t-test in Microsoft Office Excel(Microsoft,Redmond,WA,USA).
ACKNOWLEDGEMENTS
We thank Professor Dr.Eckhard Boles of Johann Wolfgang Goethe-Universität Frankfurt for providing yeast strains EBY.VW4000 and plasmid.This work was supported by grants from the National Program of High Technology Development of China(2012AA10A303),the National Natural Science Foundation of China(J1103510),and the Fundamental Research Funds for the Central Universities(2011QC068).
Altschul SF,Madden TL,Schaffer AA,Zhang J,Zhang Z,Miller W,Lipman DJ(1997)Gapped BLAST andPSI-BLAST:A new generationofprotein database search programs.Nucleic Acids Res 25:3389–3402
Ammiraju JS,Luo M,Goicoechea JL,Wang W,Kudrna D,Mueller C,Talag J,Kim H,Sisneros NB,Blackmon B,Fang E,Tomkins JB,Brar D,Mackill D,McCouch S,Kurata N,Lambert G,Galbraith DW,Arumuganathan K,Rao K,Walling JG,Gill N,Yu Y,SanMiguel P,Soderlund C,Jackson S,Wing RA(2006)The Oryza bacterial artificial chromosome library resource:Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza.Genome Res 16:140–147
Ammiraju JS,Lu F,Sanyal A,Yu Y,Song X,Jiang N,Pontaroli AC,Rambo T,Currie J,Collura K,Talag J,Fan C,Goicoechea JL,Zuccolo A,Chen J,Bennetzen JL,Chen M,Jackson S,Wing RA(2008)Dynamic evolution of oryza genome is revealed by comparative genomic analysis of a genus-wide vertical data set.Plant Cell 20:3191–3209
Antony G,Zhou J,Huang S,Li T,Liu B,White F,Yang B(2010)Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3.Plant Cell 22:3864–3876
Artero RD,Terol-Alcayde J,Paricio N,Ring J,Barques M,Torres A,Perez-Alonso M(1998)Saliva,a new Drosophila gene expressed in the embryonic salivary glands with homologues in plants and vertebrates.Mech Dev 75:159–162
Bari R,Jones JD(2009)Role of plant hormones in plant defence responses.Plant Mol Biol 69:473–488
Chen LQ,Hou BH,Lalonde S,Takanaga H,Hartung ML,Qu XQ,Guo WJ,Kim JG,Underwood W,Chaudhuri B,Chermak D,Antony G,White FF,Somerville SC,Mudgett MB,Frommer WB(2010)Sugar transporters for intercellular exchange and nutrition of pathogens.Nature 468:527–532
Chen LQ,Qu XQ,Hou BH,Sosso D,Osorio S,Fernie AR,Frommer WB(2012)Sucrose efflux mediated by SWEET proteins as a key step for phloem transport.Science 335:207–211
Chu Z,Yuan M,Yao J,Ge X,Yuan B,Xu C,Li X,Fu B,Li Z,Bennetzen JL,Zhang Q,Wang S(2006)Promoter mutations of an essential gene for pollen development result in disease resistance in rice.Genes Dev 20:1250–1255
Gamas P,Niebel Fde C,Lescure N,Cullimore J(1996)Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development.Mol Plant Microbe Interact 9:233–242
Ge S,Sang T,Lu BR,Hong DY(1996)Phylogeny of rice genomes with emphasis on origins of allotetraploid species.Proc Natl Acad Sci USA 96:14400–14405
Guindon S,Dufayard JF,Lefort V,Anisimova M,Hordijk W,Gascuel O(2010)New algorithms and methods to estimate maximumlikelihood phylogenies:Assessing the performance of PhyML 3.0.Syst Biol 59:307–321
Hamacher T,Becker J,Gardonyi M,Hahn-Hagerdal B,Boles E(2002)Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization.Microbiology 148:2783–2788
Ito H,Fukada Y,Murata K,Fields S(1983)Transformation of intact yeast cells treated with alkali cations.J Bacteriol 153:163–168
Khush GS(1997)Origin,dispersal,cultivation and variation of rice.Plant Mol Biol 35:25–34
Kim H,San Miguel P,Nelson W,Collura K,Wissptski M,Walling JG,Kim JP,Jackson SA,Soderlund C,Wing RA(2007)Comparative physical mapping between Oryza sativa(AA genome type)and O.punctata(BB genome type).Genetics 176:379–390
Kong Z,Li M,Yang W,Xu W,Xue Y(2006)A novel nuclear-localized CCCH-type zinc finger protein,OsDOS,in involved in delaying leaf senescence in rice.Plant Physiol 141:1376–1388
Lee RH,Wang CH,Huang LT,Chen SC(2001)Leaf senescence in rice plants:Cloning and characterization of senescence up-regulated genes.J Exp Bot 52:1117–1121
Li T,Huang S,Zhou J,Yang B(2013)Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv.oryzae in rice.Mol Plant 6:781–789
Lim PO,Kim HJ,Nam HG(2007)Leaf senescence.Annu Rev Plant Biol 58:115–136
Liu Q,Yuan M,Zhou Y,Li X,Xiao J,Wang S(2011)A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice.Plant Cell Environ 34:1958–1969
Mumberg D,Muller R,Funk M(1995)Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds.Gene 156:119–122
Nose Y,Rees EM,Thiele DJ(2006)Structure of the Ctr1 copper trans’PORE’ter reveals novel architecture.Trends Biochem Sci 31:604–607
Qiu D,Xiao J,Ding X,Xiong M,Cai M,Cao Y,Li X,Xu C,Wang S(2007)OsWRKY13 mediates rice disease resistance by regulating defenserelated genes in salicylate-and jasmonate-dependent signaling.Mol Plant Microbe Interact 20:492–499
Quirino BF,Normanly J,Amasino RM(1999)Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes.Plant Mol Biol 40:267–278
Seo PJ,Park JM,Kang SK,Kim SG,Park CM(2011)An Arabidopsis senescence-associated SAG29 regulates cell viability under high salinity.Planta 233:189–200
Talavera G,Castresana J(2007)Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments.Syst Biol 56:564–577
Tamura K,Peterson D,Peterson N,Stecher G,Nei M,Kumar S(2011)MEGA 5:Molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimony methods.Mol Biol Evol 28:2731–2739
Wang S,Liu N,Peng K,Zhang Q(1999)The distribution and copy number of copia-like retrotransposons in rice(Oryza sativa L.)and their implications in the organization and evolution of the rice genome.Proc Natl Acad Sci USA 96:6824–6828
Wang S,Liu K,Zhang Q(2000)Segmental duplications are common in rice genome.Acta Bot Sin 42:1150–1155
Wang L,Xie W,Chen Y,Tang W,Yang J,Ye R,Liu L,Xu C,Xiao J,Zhang Q(2010)A dynamic gene expression atlas covering the entire life cycle of rice.Plant J 61:752–766
Wieczorke R,Krampe S,Weierstall T,Freidel K,Hollenberg CP,Boles E(1999)Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae.FEBS Lett 464:123–128
Wintermans JF,de Mots A(1965)Spectrophometric characteristics of chlorophylls a and b and their phenophytins in ethanol.Biochim Biophys Acta 109:448–453
Xiao W,Liu H,Li Y,Li X,Xu C,Long M,Wang S(2009)A rice gene of de novo origin negatively regulates pathogen-induced defense response.PLoS ONE 4:e4603
Yang B,Sugio A,White FF(2006)Os8N3 is a host disease-susceptibility gene for bacterial blight of rice.Proc Natl Acad Sci USA 103:10503–10508
Ye L,Berden JA,van Dam K,Kruckeberg AL(2001)Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiae.Yeast 18:1257–1267
Yuan M,Wang S(2013)Rice MtN3/saliva/SWEET family genes and their homologues in cellular organisms.Mol Plant 6:665–674
Yuan M,Chu Z,Li X,Xu C,Wang S(2010)The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution.Plant Cell 22:3164–3176
Yuan M,Li X,Xiao J,Wang S(2011)Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice.BMC Plant Biol 11:69
Zhou B,Peng K,Chu Z,Wang S,Zhang Q(2002)The defense-responsive genes showing enhanced and repressed expression after pathogen infection in rice(Oryza sativa L.).Sci China C Life Sci 45:449–467
SUPPORTING INFORMATION
Additional supporting information can be found in the online version of this article:
Figure S1.The structure of rice MtN3/saliva/SWEET-type genes UTR,untranslated region
Figure S2.Phylogenetic relationship of rice MtN3/saliva/SWEET proteins analyzed by maximum parsimony algorithm method(A)and maximum likelihood method(B)
Figure S3.The alignment of highly conserved regions of rice MtN3/saliva/SWEET protein sequences
Figure S4.Phylogenetic relationship of rice MtN3/saliva/SWEET proteins analyzed by neighbor-joining method(A),maximum likelihood method(B),and maximum parsimony algorithm method(C)
Figure S5.Expression profiles of MtN3/saliva/SWEET paralogs in rice seedlings after phytohormone treatment
Table S1.MtN3/saliva/SWEET-type genes in rice genome
Table S2.Wild rice materials
Table S3.Polymerase chain reaction(PCR)primers used for genomic DNA amplification MtN3/saliva/SWEET-type genes
Table S4.Polymerase chain reaction(PCR)primers used for reverse transcription(RT)-PCR or quantitative q(RT)-PCR assays
Table S5.Polymerase chain reaction(PCR)primers used for yeast complementation experiments
 Journal of Integrative Plant Biology2014年6期
Journal of Integrative Plant Biology2014年6期
- Journal of Integrative Plant Biology的其它文章
- A step-by-step protocol for formaldehyde-assisted isolation of regulatory elements from Arabidopsis thaliana
- A new loss-of-function allele 28y reveals a role of ARGONAUTE1 in limiting asymmetric division of stomatal lineage ground cell
- Polycomb-group histone methyltransferase CLF is required for proper somatic recombination in Arabidopsis
- Molecular characterization and expression analysis of Triticum aestivum squamosa-promoter binding protein-box genes involved in ear development
- BnWRI1 coordinates fatty acid biosynthesis and photosynthesis pathways during oil accumulation in rapeseed
- Genetic analysis of biomass and photosynthetic parameters in wheat grown in different light intensities
