Genetic variations in triple-negative breast cancers undergoing neo-adjuvant chemotherapy
Miki Mori,Tomoko Watanabe,Sadako Akashi-Tanaka,Kumiko Ueda,Reiko Makino,Yuko Hirota,Seigo Nakamura
1Department of Breast Surgical Oncology,Showa University,Shinagawaku,Tokyo 142-8666,Japan.
2Clinical Research Laboratory,Showa University,Shinagawaku,Tokyo 142-8666,Japan.
3Department of Pathology,Showa University,Shinagawaku,Tokyo 142-8666,Japan.
Abstract
Aim:Triple negative breast cancer (TNBC) is known as aggressive subtype and have no identified targeted therapies.We examined the relationship of neoadjuvant chemotherapy response to genetic variations of TNBC.
Methods: The tumors used in this study were collected from Showa University Hospital,Japan.Thirteen formalinfixed paraffin-embedded tumors from Japanese TNBC patients who underwent neoadjuvant chemotherapy were used for analysis.Of these,eight surgically resected tumors showed progressive disease and/or recurrence after treatment (PD/REC),and biopsy tissues from five patients showing pathological complete response (pCR) were analyzed.DNA extracted from tissue sample were analyzed.The Miseq system and Trusight Tumor Sequence panel kit were used to sequence 174 amplicons over 82 exons of 26 cancer-related genes to identify genetic mutations.
Results:Seven somatic non-synonymous variants were detected in three genes (FOXL2,PIK3CA,and TP53) in all five pCR patients,and six somatic non-synonymous variants in two genes (PTEN and TP53) were detected in six of eight PD/REC patients.Eight of 13 TNBC tumors were found to have TP53 pathogenic variants,in both pCR and PD/REC cases.
Conclusion:Although TP53 variation was detected in both pCR and PD/REC cases,each location and type of the variant were different.We could not identify genetic mutations associated with chemotherapy response and recurrence.
Keywords: Triple negative breast cancer,next generation sequence,TP53 and neo adjuvant chemotherapy
INTRODUCTION
Triple-negative breast cancer (TNBC) is defined as any breast cancer lacking estrogen receptor,progesterone receptor,and human epidermal growth factor receptor 2 expression.It accounts for approximately 15%-20% of all breast cancers[1,2].TNBC is reported to behave more aggressively and is associated with a worse survival than other types of breast cancer[3,4].Patients with TNBC have no identified targeted therapies.They are currently treated by surgery and chemotherapy alone[1].
Triple-negative subtypes are more sensitive to neo-adjuvant chemotherapy than luminal breast cancers[5-7].Patients with TNBC have increased pathologic complete response (pCR) rates compared with non-TNBC patients,and those with pCR have excellent survival regardless of subtype[5-7].However,patients with progressive disease after neo-adjuvant chemotherapy have a significantly worse prognosis if they have TNBC compared with other subtypes[5,7].About 70%-80% of patients of TNBC fail to respond to neoadjuvant chemotherapy (NAC)[5-7],but the molecular mechanism underlying differences in responders and non-responders is unclear.It is also poorly understood which genes and pathways are related to treatment response in breast tumors.
Herein,we performed an exploratory study using next generation sequencing to investigate the dynamics of tumor gene mutations in TNBC patients receiving NAC.
METHODS
Ethical approval
This study was approved (number 1589) by the Institutional Review Board of Showa University,Japan.Written informed consent was received from the patients.If informed consent could not be received for the death of patients,the candidate opted out on the homepage of the hospital.
Patient information
The tumors analyzed in this study were collected from Showa University Hospital,Japan.Ninety four patients revealed triple negative subtype of 1111 patients of primary breast cancer surgery between 2011 and 2013.Of these 94 patients,33 underwent neo adjuvant chemotherapy prior to surgery.Ten patients achieved pCR and remaining 23 patients had residual disease.Four patients revealed clinically progressive disease during neo adjuvant chemotherapy.Taxane followed by anthracycline based regimens were applied as standard neo-adjuvant chemotherapy.Two patients who revealed progressive disease during first taxane regimen dropped out of neo-adjuvant chemotherapy then planned surgery without following anthracycline chemotherapy.Seven patients of 23 patients who had residual disease including 3 progressive disease revealed recurrence in the same period.In total,there were 8 patients with progressive disease and/or recurrence (PD/REC).Five of 10 pCR cases underwent pretreatment biopsy in Showa University Hospital and able to obtain the sample.These 8 PD/REC cases and 5 pCR cases were analyzed.Formalinfixed paraffin-embedded (FFPE) samples from pretreatment biopsy of PD/REC and pCR cases and surgical specimen of PD/REC cases were obtained.Clinical and pathologic information was obtained from medical chart including the information of the results of both germline BRCA1/2 testing and BRNAness.The concept of “BRCAness” is that sporadic basal-like breast cancers resemble BRCA1 mutated cancers.A set of 34 MLPA probes was used to identify BRCAness[8].American Joint Committee on Cancer/tumor-nodemetastasis cancer staging was assessed.
Preparation of genomic DNA
Ten slices of 10 μm-thick FFPE tissues were used for genomic DNA extraction.Tissues were de-paraffinized with xylene,then DNA was extracted using the QIAamPD/recNA FFPE Tissue Kit (Qiagen,Venlo,Netherlands) following the manufacturer's instructions.
Briefly,each deparaffinization solution,specific buffers and proteinase was added to destruct cells while centrifuging.After incubation,ethanol was added to precipitate DNA.After transferring the entire lysate to purification column,the centrifugation was repeated.Finally,elution buffer was added to elute DNA in the column.
Target genes
The TruSight Tumor Sequencing panel (Illumina,San Diego,California,USA,#FC-130-2001) was used to detect genetic variations in tumors using NGS.This panel covered 174 amplicons over 82 exons of 26 cancer-related genes.The following genes encode kinases:AKT1,ALK,BRAF,EGFR,ERBB2,FGFR2,KIT,MAP2K1,MET,PD/RECGFRA,PIK3CA,SRC,andSTK11,while the remainder do not:APC,CDH1,CTNNB1,FBXW7,FOXL2,GNAQ,GNAS,KRAS,MSH6,NRAS,PTEN,SMAD4,andTP53.
Sequencing
The quality of the genomic DNA purified from FFPE tissues was checked by quantitative PCR using Quality Control Template (Illumina) and iQTMSYBR® Green Supermix (BioRad).Samples were required to fulfill the criteria ΔCt < 5,which was typically 30-300 ng depending on the DNA quality.However,ΔCt = 12 biopsy specimens were also included in the analysis.Genomic DNA hybridization-based enrichment was performed using the TruSight Tumor Sequence panel kit according to the manufacturer's protocol.Pooled amplicons were end-repaired then underwent adapter ligation.The purified library was quantified using the Qubit® fluorometer with the Qubit® dsDNA BR assay kit.Finally,the captured DNA was subjected to 121-bp paired-end read sequencing on the MiSeq system (Illumina).
Strategies for the analysis of genetic variations
Sequence reads were exported into FASTQ format files.The alignment of pair-end sequencing reads to the reference genome was performed using a banded Smith-Waterman method with the Amplicon-DS workflow software plug-in for MiSeq Reporter.Human Genome version 19 (UCSC) was used as the alignment reference.The identification of sequencing variants was performed using the Illumina Somatic Variant Caller algorithm.The called SNPs/indels were annotated using COSMIC (somatic mutation information),the IARC TP53 database (human TP53 gene variations related to cancer)[9],and other databases of genetic variations.Database was aggregated into the Illumina VariantStudio application.Additionally,NM_000179.2:c.3254delC and NM_000179.2:c.3253_3254insC inMSH6were eliminated as known artifact variants.Statistical analysis was performed with Fisher's exact test.
RESULTS
Clinical features of the patients
The clinical features of the patients are summarized in the Table 1.Patients had an average age of 49.6 years.Seven cases underwentBRCA1andBRCA2germline mutation testing.BRCA1/2gene testing are not covered by national healthcare insurance,so it was performed to limited patients based on age,family history by self-pay.Seven PD/REC cases,including three patients with germline mutation showed BRCAness.One pCR case also showed BRCAness.
Genetic variations in tumors showed PD/REC and pCR
The genetic variations after filtering out synonymous variants and known artifacts are shown in the Table 2.Three biopsy samples of 8 PD/REC cases were not available for the analysis.In PD/REC cases,we found ten non-synonymous variants were detected in the coding regions of two genes in 5 biopsy tissues before NAC,while 14 non-synonymous variants were detected in the coding regions of three genes in 8 surgical specimens after NAC.In 5 pCR cases,ten non-synonymous variants were detected in the coding regions of four genes.Of these,variantsTP53Val31Ile and Pro72Arg,STK11Phe354Leu,andMETAsn375Ser are known polymorphisms (NIH dbSNP:rs33917957,rs1042522,rs59912467 and rs33917957,respectively).

Table 1.Clinical features of the patients
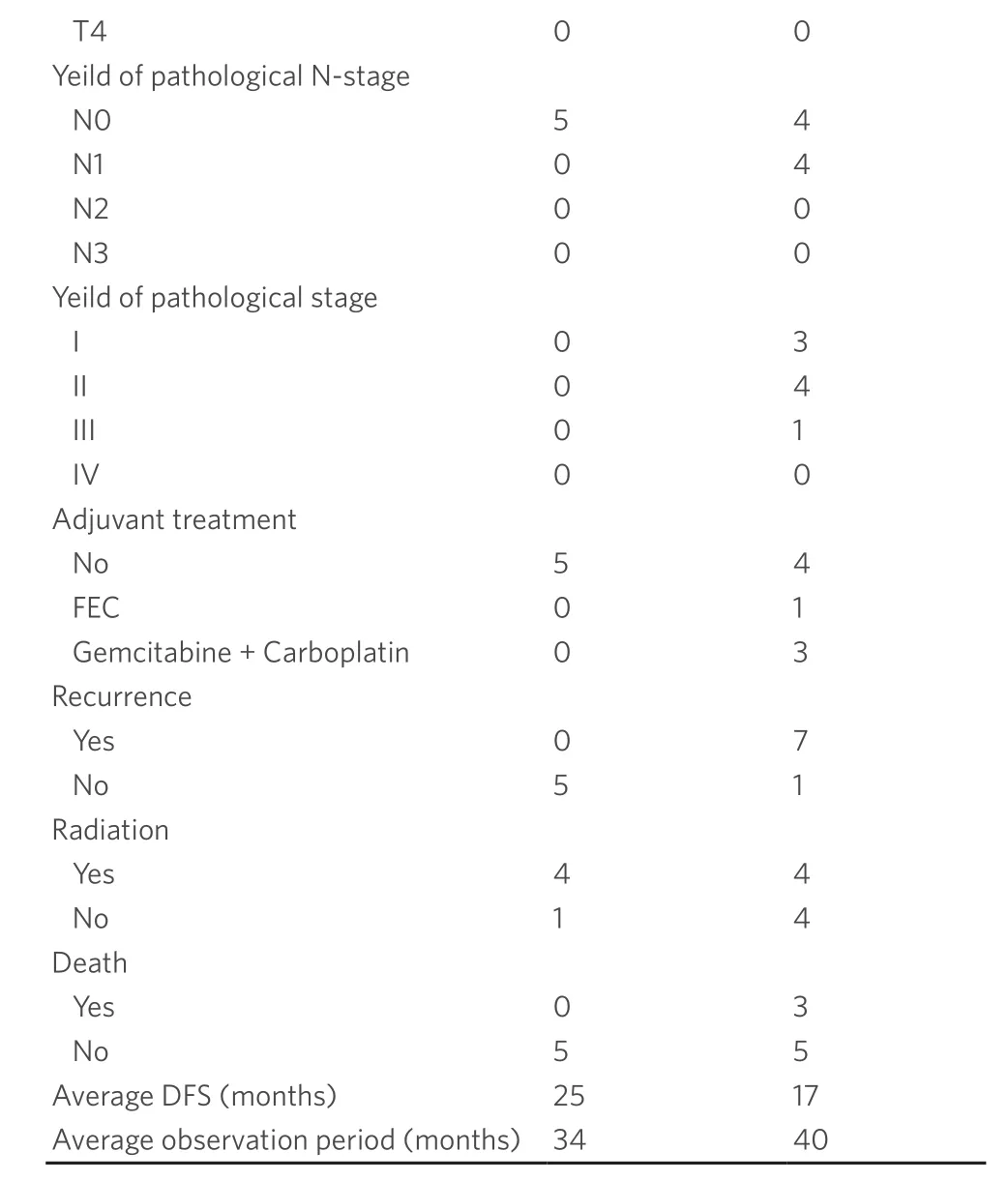
pCR:pathological complete response; PD/REC:progressive disease and/or recurrence; IHC:immunohistochemistry; DFS:disease free survival; FEC:fluorouracil epirubicin cyclophosphamide
As a result,total of 3 somatic mutations in one gene (TP53) and six somatic mutations in two genes (TP53andPTEN) were detected in biopsy samples (3/5 cases) and surgical specimens of PD/REC patients (6/8 cases),respectively.Somatic mutations were detected in three genesTP53,FOXL2andPIK3CAin pCR patients (5/5 cases).
TP53was the most frequently mutated gene (9 of 13,69.2% in total,5 of 8,62.5% in PD/REC cases and 4 of 5,80% in pCR cases) (P= 0.50).Other pathogenic variants detected at a lower frequency were in genesPIK3CA(n= 1),PTEN(n= 1) andFOXL2(n= 1).In both PD/REC and CR,unique pathogenic variants were detected inTP53and other genes.As shown in the Table 3,theTP53Pro72Arg polymorphism was detected in both pCR and PD/REC cases.
Next,we compared the effect of chemotherapy in five matched cases showed PD/REC.In patient 7,thePTENSer179Valfs★8 mutation was detected after NAC,but it was seen before NAC at low frequency on one strand (2%).Other genetic variations (TP53andSTK11) were observed both pre- and post-treatment.
DISCUSSION
In this study,we used NGS to analyze 26 genes of 13 TNBC tumors receiving NAC.This is the first report to determine genetic variations in Japanese TNBC patients who showed pCR and PD/REC as a result of NAC.TP53pathogenic variants were detected in both pCR and PD/REC cases at frequencies similar to the high rates previously reported.TP53genetic variations in pCR cases might be associated with sensitivity to chemotherapy,and otherTP53variants in PD/REC cases might affect resistance to chemotherapy.Thus,sensitivity to chemotherapy may vary by the location and type of somaticTP53variants.However,we could not confirm whether these genetic alterations found in this study were responsible for the chemosensitivity and recurrence because the number of patients are too small.

Table 2.Genetic vatiations detected in pCR/PD groups
The numbers of genes and cases assessed were limited in this study.However,the frequency of genetic variations detected in TNBC did not differ greatly from those in previous studies.Lipset al.[10]analyzed 1977 genes in 56 pre-treatment TNBC biopsies of NAC responders and non-responders as well as matched normal DNA.They reported that most mutations were inTP53,TTN,andPIK3CA(55%,14%,and 9%,respectively).No recurrent mutations were associated with chemotherapy response or relapse[10].Balkoet al.[11]examined residual disease in 74 clinically defined TNBCs after NAC,which included NGS performed on 20 matched pretreatment biopsies.Targeted NGS was carried out of 3320 exons from 182 oncogenes and tumor suppressors as well as 37 introns of 14 genes frequently rearranged in cancer.The frequency of each mutation was fewer than 5% of samples.Alterations inTP53were identified in 72 of 81 samples (89%),which was a similar rate to that observed in other studies of basal-like breast cancer or TNBC,and The Cancer Genome Atlas dataset (~85%)[11].Roy-Chowdhuriet al.[12]reported that a triple-negative group (n= 77) showed mutations in 15 of 46 genes tested,withTP53showing the highest mutation frequency (n= 48/77,62%),followed byPIK3CA(n= 13/77,17%),APC,RET,SMAD4(n= 2,3%),AKT1,ATM,BRAF,FGFR1,HRAS,JAK3,MET,SRC,PTEN,andSTK11(n= 1,1%)[12].From these previous reports,it is difficult to conclude that the varied and sparse genetic alterations seen in the present study caused differences between responders and non-responders toward chemotherapy.
TP53mutations in breast cancer have previously been reported to be associated with worse prognosis[13,14].Ooeet al.[15]investigated the relationship between the p53 mutation status and response to docetaxel in breast cancers.They found that the response rate of patients with p53-mutated tumors (44%) was lower than that of those with wild-type tumors (62%).In addition,theTP53p.Pro72Arg polymorphism was reported association with platinum-based chemotherapy response in non-small cell lung cancer[16].Platinum-based chemotherapy was only administered to one case in our study,so no response difference was detected between heterozygous and homozygous variants in the pCR or PD/REC group.However,the same genetic variations ofTP53(p.V157F and p.Cys141Tyr) detected in PD/REC samples in previous study[11]were also observed in pCR samples in our study.

Table 3.Genotype of the TP53 Pro72Arg polymorphism
As a cohort,it is representative of Japanese TNBC,but the number of patients and samples was too low to confirm relevant relationship with genetic variants.This preliminary study should be followed up with a higher number of patients and selected genes in future.
In this study,8 of 13 TNBC tumors were found to haveTP53pathogenic variants,in both pCR and PD/REC cases.TP53variations were detected at similarly high rates as previously reported and may be associated with sensitivity or resistance to chemotherapy depending on the location and type of the variant.If the effect of these genetic variations on the induction of apoptosis could be determined,this might enable the mechanism of the NAC chemotherapy response to be understood.Our results provide insights into potentially actionable variants for targeted therapeutic options in TNBC.
DECLARATIONS
Authors' contributions
Drafting of the manuscript,review and revisions of the final draft,data analysis and experiments:Mori M,Watanabe T,Makino R
Experiments:Ueda K,Hirota Y
Supervision,manuscript review and project administration:Akashi-Tanaka S,Nakamura S
Availability of data and materials
Not applicable.
Financial support and sponsorship
This work was supported in part by a Grant-in-Aid for the High-Technology Research Center Project from the Ministry of Education,Culture,Sports,Science and Technology of Japan.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
This study was approved (number 1589) by the Institutional Review Board of Showa University,Japan.Written informed consent was received from the patients.If informed consent could not be received for the death of patients,the candidate opted out on the homepage of the hospital.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2019.
- Cancer Drug Resistance的其它文章
- Enhanced Kat3A/Catenin transcription:a common mechanism of therapeutic resistance
- Cancer drug resistance:rationale for drug delivery systems and targeted inhibition of HSP90 family proteins
- Circulating non-coding RNAs in recurrent and metastatic ovarian cancer
- Computational analyses for cancer biology based on exhaustive experimental backgrounds
- Dodging the bullet:therapeutic resistance mechanisms in pediatric cancers
- Drug-adapted cancer cell lines as preclinical models of acquired resistance