Gene testing for osteonecrosis of the femoral head in systemic lupus erythematosus using targeted next-generation sequencing:A pilot study
Hong-Sheng Sun,Qing-Rui Yang,Yan-Yan Bai,Nai-Wen Hu,Dong-Xia Liu,Department of Rheumatology and Immunology,Shandong Provincial Hospital Affiliated to Shandong University,Jinan 250021,Shandong Province,China
Cheng-Yong Qin,Department of Gastroenterology,Shandong Provincial Hospital Affiliated to Shandong University,Jinan 250021,Shandong Province,China
Abstract
Key words:Single nucleotide variations;Osteonecrosis of the femoral head;Systemic lupus erythematosus;Nitric oxide synthase 3;Collagen type II alpha 1 chain;Complement C3d receptor 2
INTRODUCTION
Both osteonecrosis of the femoral head (ONFH) and systemic lupus erythematosus(SLE) are multifactorial and complex diseases caused by both genetic alterations and environmental exposures[1].Risk factors for secondary ONFH are known to involve the intake of corticosteroids,autoimmune disease,alcohol abuse,and radiation.Epidemiological studies suggest that SLE is the most common autoimmune disease to be the primary cause of steroidinduced ONFH[2].However,only some patients with SLE receiving steroid administration ultimately develop ONFH,and there are a number of patients with SLE with ONFH who have no experience of corticosteroid treatment[3].These reports indicated that other reasons,such as genetic variations between individuals,might also be involved in the onset of ONFH in SLE.Indeed,gene polymorphisms that affect coagulation,metabolic factors,mechanical stresses,immunologic factors,and fibrinolytic systems have been identified[4,5]and some of these genes have been suggested to be involved in SLE with ONFH.
Complement receptor type 2 (CR2) is a transmembrane glycoprotein expressed in mature B and follicular dendritic cells.CR2 plays a role in complement activation,antigen targeting,and B cell activation.Endothelial nitric oxide synthase (NOS3)regulates bone formation and osteoblast function.It has been reported that in Korean patients with SLE,CR2 and NOS3 contribute to ONFH susceptibility[6,7].Collagen type II alpha-1 gene (COL2A1) is a causal gene of skeletal dysplasia and epiphyseal dysplasia.COL2A1mutations cause familial idiopathic ONFH[8];however,the relationship was not found in Japanese idiopathic ONFH patients[9].PTPN22encodes protein tyrosine phosphatase nonreceptor 22,a lymphoid protein,mutations in which might promote T cell activation and thus cause autoimmune diseases,such as rheumatoid arthritis,SLE,and juvenile idiopathic arthritis[10,11].Transient receptor potential vanilloid 4 (TRPV4) forms a calciumpermeable non-selective cation channel and plays a role in vasoregulation and osteoclast differentiation.A novelTRPV4mutation and altered calcium homeostasis have been observed in ONFH[12].
In this study,we investigated whether patients with SLE with ONFH have a genetic predisposition to ONFH in SLE,the identification of which might lead to more efficient diagnosis,evaluation,and even prevention of the disease.Using FastTarget and Illumina Miseq sequencing technologies,we analyzed single nucleotide variations (SNVs) inCR2,NOS3,COL2A1,PTPN22,andTRPV4genes of patients with SLE with ONFH.
MATERIALS AND METHODS
Patients included in the study
We enrolled 49 patients with SLE with ONFH (4 males and 45 females;mean age:33.57 ± 11.24 years) visiting the Department of Rheumatology and Immunology of Shandong Provincial Hospital Affiliated to Shandong University.All patients met the criteria of the American College of Rheumatology as revised in 1997[13].Individuals provided samples of peripheral blood,2 mL of which was used for genomic DNA isolation using a DNeasy Blood &Tissue Kit (QIAGEN,Hilden,Germany) following the manufacturer's standard procedure.The local Ethics Review Board approved the current study,and informed consent was provided by all participants.
Mutation analysis
Primers were designed using Primer 3.To cover all the coding sequences and most of the untranslated regions of the five genes that might be responsible for ONFH in SLE,243 oligonucleotide pairs were produced.A first round of primer design using the most stringent conditions (e.g.,no single nucleotide polymorphisms in the primer annealing region,an amplicon comprising 200-270 bp,and a GC content of 30%-80%)allowed us to put the 243 oligonucleotide pairs into 15 multiplex PCR panels to amplify the target regions from the five genes.An ABI 2720 Thermal Cycler (Life Technologies Corporation,Carlsbad,CA,United States) was used to carry out the amplification reactions using following cycling program:95°C for 2 min;11 cycles of 94 °C for 20 s,63.5 °C for 40 s,and 72 °C for 1 min;24 cycles of 94 °C for 20 s,65 °C for 30 s,and 72 °C for 1 min;and final incubation at 72 °C for 2 min.An 8 bp barcode was incorporated into each PCR product,and then all the libraries for each sample were pooled.Cluster generation and hybridization of the sequencing primers were followed by nucleotide sequencing carried out on a MiSeq Benchtop Sequencer(Illumina,Inc,San Diego,CA,United States) in one single lane,following the manufacturer's standard protocols.For each sequencing read,300 cycles were carried out to produce paired-end reads including 300 bp at each end and the 8-bp index tag.
Bioinformatic analysis
The Burrows-wheeler aligner[14]method was used to align the sequencing reads to human genome version hg19.The GATK[15]and Varscan programs[16]were used to call the SNVs,the data for which were then combined.SNVs were annotated using the Annovar program[17].PolyPhen-2,SIFT,and MutationTaster[18-20]were used to assess the functional effects of non-synonymous SNVs.Significantly non-benign nonsynonymous SNVs were identified as those with a Polyphen-2 score of>0.85,a SIFT score of < 0.05,or a MutationTaster score of>0.85.Perl scripts then filtered the SNVs against those of dbSNP135 to segregate benign polymorphisms from potentially deleterious variants.Benign polymorphisms were considered as those SNVs present in dbSNP135 with a minor allele frequency of ≥ 1% in a Chinese population from the 1000 genome database,and were removed from subsequent analysis.
Statistical analysis
The two-tailed Student’st-test and Fisher’s exact test orχ2test were used when appropriate.Differences were considered significant if thePvalue was less than 0.05.The odds ratio and 95% confidence interval (95%CI) were calculated.
RESULTS
SNV identification in patients with SLE with ONFH
To identify SNVs,VarScan (http://varscan.sourceforge.net/) a nd GATK HaplotypeCaller (https://software.broadinstitute.org/gatk/best-practices//) were used to analyze single nucleotide polymorphisms and insertion-deletions (InDels) in the samples.A total of 92 SNVs and 17 InDels were identified using both approaches,with 4 SNVs and 1 InDel being identified only by VarScan,and another 16 SNVs and 2 InDels only by GATK HaplotypeCaller.
The identified SNVs and InDels were then grouped by the corresponding regions of the genome,including:Intron (40.15%),exon (21.21%),intergenic (13.64%),ncRNA(non-coding)_intron (7.58%),3′ untranslated region (UTR;5.3%),splicing site (10 bp around a splice junction) (4.55%),upstream (1 kb upstream of the transcription start site;3.03%);downstream (between one gene’s upstream and another gene’s downstream;1.52%),5′ UTR (1.52%),exonic splicing (0.76%),and ncRNA (noncoding)_splicing (0.76%).In this study,15 SNVs were nonsynonymous (51.72%),while the other 14 SNVs were synonymous (48.28%).Gene transition (Ts) and transvertion (Tv) (both as annotated by dbSNP) of the SNVs were also analyzed.There were 61 (54.46%) Ts events and 29 (25.89%) Tv events,giving a Ts:Tv ratio of 2.1034.There were 15 (13.39%) Novel_Ts events (not annotated by dbSNP) and 7(6.25%) Novel_Tv events (not annotated by dbSNP),giving a Novel_Ts:Tv ratio of 2.1429.
For all the identified InDels,2 insertions comprised 1-5 bp (10%),while 17 deletions comprised 1-5 bp (85%) and 1 deletion was 6-10 bp (5%).The genomic distributions of the SNVs for each sample are shown in Table1.
Low frequency SNVs in patients with SLE with ONFH
Among the 49 patients,six (12.25%) were confirmed to have low frequency functional mutations,including one patient with a mutation in theNOS3gene,four patients with a mutation in theCOL2A1gene,and one patient with a mutation in theCR2gene;and the first priority (Take the highest priority of SNVs if the mutation is dominant or homozygous;take the lower one from the top two highest priority SNVs if it is a heterozygous recessive pattern) of these gene mutations were third,first1,and second,respectively.However,we failed to detect significant differences in the frequency of these mutations in comparison with controls (Table2).
The low frequency functional mutations inNOS3are shown as follows:NM_001160109:exon 6:c.814G>A:p.E272K,NOS3:NM_000603:exon 7:c.814G>A:p.E272K,NOS3:NM_001160110:exon 6:c.814G>A:p.E272K,and NOS3:NM_001160111:exon 6:c.814G>A:p.E272K (Figure1),which had never been reported previously.Most of these mutations here comprised nonsynonymous SNVs and were predicted as tolerated (SIFT Score Pred),possibly damaging (POLYPHEN Score Pred),and disease_causing damaging (MutationTaster Score Pred) (Table3).
We revealed two rare functional mutations ofCOL2A1,both of which are nonsynonymous SNVs.One is rs41263847:COL2A1:NM_001844:exon 29:c.1913C>T:p.T638I;COL2A1:NM_033150:exon 28:c.1706C>T:p.T569I,the first priority of which is second;and was predicted as tolerated (SIFT Score Pred),benign (POLYPHEN Score Pred),and disease_causing (MutationTaster Score Pred ) damaging.The other is rs371445823 COL2A1:NM_001844:exon 8:c.580G>A:p.A194T,COL2A1:NM_033150:exon 7:c.373G>A:p.A125T (Figure2),the first priority of which is first1;and was predicted as tolerated (SIFT Score Pred),possibly damaging (POLYPHEN Score Pred),and disease_causing damaging (MutationTaster Score Pred ) (Table3).
One SNV identified in theCR2gene is also a nonsynonymous SNV with the first priority being second:rs45573035:CR2:NM_001006658:exon 2:c.200C>G:p.T67S,CR2:NM_001877:exon 2:c.200C>G:p.T67S (Figure3).The variations were predicted as tolerated (SIFT Score Pred),possibly damaging (POLYPHEN Score Pred),and polymorphism damaging (MutationTaster Score Pred) (Table3).
The phenotypic features of patients with SLE with ONFH are listed in Table4.The patient with mutations inNOS3was a 36-year-old woman who had suffered from SLE for 4 years and ONFH for 1 mo,who had skin rashes and arthritis.For the patient with mutations inCR2,she also had skin rashes and arthritis,and a renal disorder.The four patients withCOL2A1mutations (G4,G6,G34,and G42) had a relatively low SLEDAI (Systemic Lupus Erythematosus Disease Activity Index) and none of them had interstitial pneumonia,renal disorders,or neurological disorders;however,three of them had arthritis and anemia.The C3 and C4 levels,and 24-hour urinary protein levels were normal in all six patients.
DISCUSSION
The known secondary risk factors for ONFH comprise rheumatic diseases,alcohol abuse,and the use of corticosteroids.Among autoimmune diseases,SLE has a higher ONFH incidence,ranging from 5% to 30%,compared with that in the general population.Moreover,the treatment of SLE deteriorated with the onset of ONFH.Details of the pathogenesis of ONFH in SLE are unclear because patients with SLE who have not taken corticosteroids also develop ONFH.To investigate whether patients with SLE with ONFH have a genetic predisposition,we used next generation sequencing technology to analyze SNVs in reported risk genes,includingCR2,NOS3,COL2A1,PTPN22,andTRPV4.Bioinformatic analyses identified 112 SNVs and 20
InDels.Most of these genomic variations were localized in coding sequence and more than the half were nonsynonymous.Almost all insertions and deletions were 1-5 bp,except for 1 deletion that was 6-10 bp.Low frequency functional mutations ofNOS3,COL2A1,andCR2were found,although the differences between the patients and controls were not significant.

Table1 Genotype distribution of single nucleotide variants
NOS3 deficiency results in impaired osteoblast function and reduced bone formation.The mutations in theNOS3identified in the present study were all previously unreported nonsynonymous SNVs.The corresponding regions of the genome were exon 6:c.814G>A:p.E272K and exon 7:c.814G>A:p.E272K.Nitric oxide,catalyzed by endothelial nitric oxide synthase (eNOS),is involved in ONFH pathogenesis by regulating angiogenesis,thrombosis,smooth muscle proliferation,and bone turnover.Excessive nitric oxide production occurs during SLE and certain other autoimmune diseases[21].A recent study in Korean patients with SLE suggested that exonicNOS3polymorphisms,such as rs1549758 (Asp258Asp;exon 6) and rs1799983 (Glu298Asp;exon 7) might increase the risk of ONFH[7].SNP Glu298Asp inNOS3exon 7 is also associated with idiopathic ONFH in Korean patients[22,23].The c.814G>A:p.E272K mutation in exons 6 and 7 of NOS3 was also predicted to alter protein function and predicted as tolerated,possibly damaging and disease_causing damaging using several online tools.Mutations affecting the N-terminal domain of NOS3 might alter its function,leading to alterations in the enzymatic activity or expression of eNOS,thus causing ONFH in SLE.
TheCOL2A1gene is 31.5 kb,comprising 54 exons that encode a protein of 1487 amino acids with a molecular mass of 134.4 kDa.Mutations inCOL2A1result in skeletal dysplasias because of failure of cartilage development and growth,which further cause epiphyseal dysplasia of the femoral head and spinal deformity.
The cause of familial idiopathic ONFH has been reported to be four types of COL2A1 mutation in six families:c.3508G>A (p.Gly1170Ser,rs121912891);c.1888G>A(p.Gly630Ser);c.2149G>A (p.Gly717Ser,rs387906558);and c.4148C>T (p.Thr1383Met,rs138498898)[24-26].We identified two rare functional mutations in theCOL2A1gene:rs41263847:exon 29:c.1913C>T:p.T638I,exon 28:c.1706C>T:p.T569I,and rs371445823:exon 8:c.580G>A:p.A194T,exon 7:c.373G>A:p.A125T.Exons 6-48 of theCOL2A1gene encode the core region in the 330-Gly-X-Y triple-helical domain.Previous studies demonstrated that genetic mutations in the triple-helical domain can cause damage to cartilage homeostasis and long bone development.The mutations identified inCOL2A1in the present study also mapped to this domain and might impair the assembly,folding,intracellular transport,or secretion of the type II collagens in patients with SLE,ultimately resulting in ONFH.
The membrane glyocprotein CR2 binds degraded C3 fragments that are generated during complement activation.In normal immunity,CR2 has many important functions and is believed to play a role in autoimmune disease development.Previous data suggested that rs1876453 inCR2affects gene regulation and decreases susceptibility to lupus[27].The nonsynonymousCR2SNP rs17615 in exon 10 (G/A,Ser639Asn) is a conserved SNV in sheep,rats,and mice,and might affect CR2 receptor function.This mutation is associated with an increased risk of ONFH in Korean patients with SLE,possibly through impairing the normal expression of CR2[6].In the present study,we found a nonsynonymous SNV inCR2:rs45573035:exon 2:c.200C>G:p.T67S.This variation was predicted to be possibly damaging and polymorphism damaging.Thus,these mutations in exon 2 might change the function of CR2 and increase the susceptibility of patients with SLE to ONFH.
With the aid of predictive bioinformatics tools,we identified four possible pathogenic variants.Even though the size of the patient group was small,we are the first to use next generation sequencing data to identify SNVs ofCR2,NOS3,COL2A1,PTPN22,andTRPV4genes in patients with SLE and ONFH.Based on bioinformatic studies,we identified mutations ofNOS3(exon 6:c.814G>A:p.E272K and exon 7:c.814G>A:p.E272K),COL2A1 (rs41263847:exon 29:c.1913C>T:p.T638I,exon 28:c.1706C>T:p.T569I,and rs371445823:exon 8:c.580G>A:p.A194T,exon 7:c.373G>A:p.A125T) andCR2(rs45573035:exon 2:c.200C>G:p.T67S) that are likely to be associated with the development of ONFH in SLE.These findings may have important pharmacogenetic implications.However,the detailed mechanisms of the associations need to be determined in further studies.
Ethics approval and consent to participate
All the procedures that involved human participants were performed according to the ethical standards of the institutional and/or national research committee and with the1964 Helsinki declaration and its later amendments or comparable ethical standards.All individual participants included in the study provided informed consent.
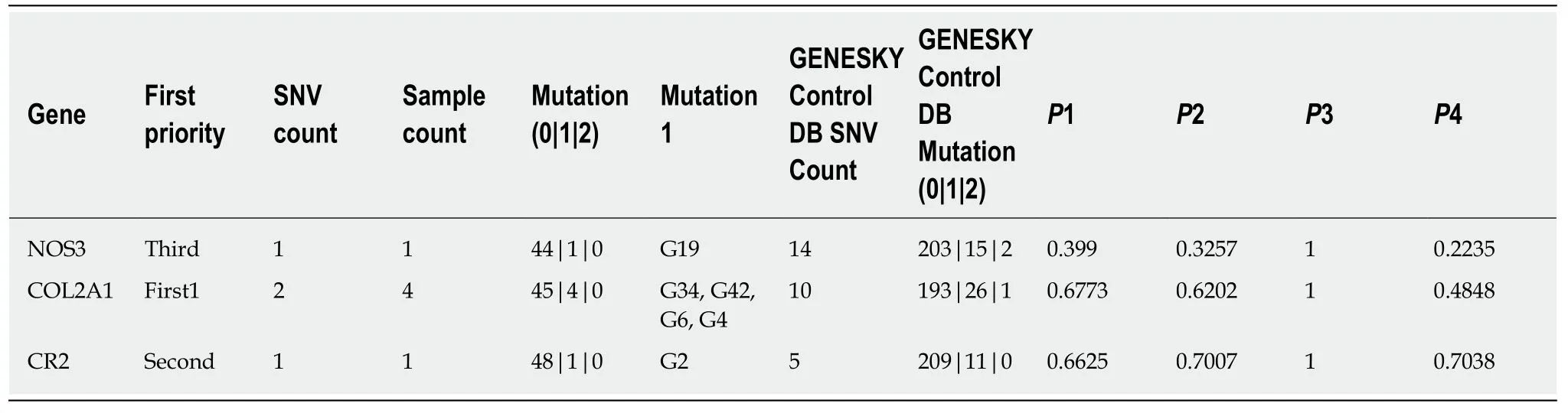
Table2 Low frequency functional mutations

Table3 Single nucleotide variant information for low frequency functional mutations
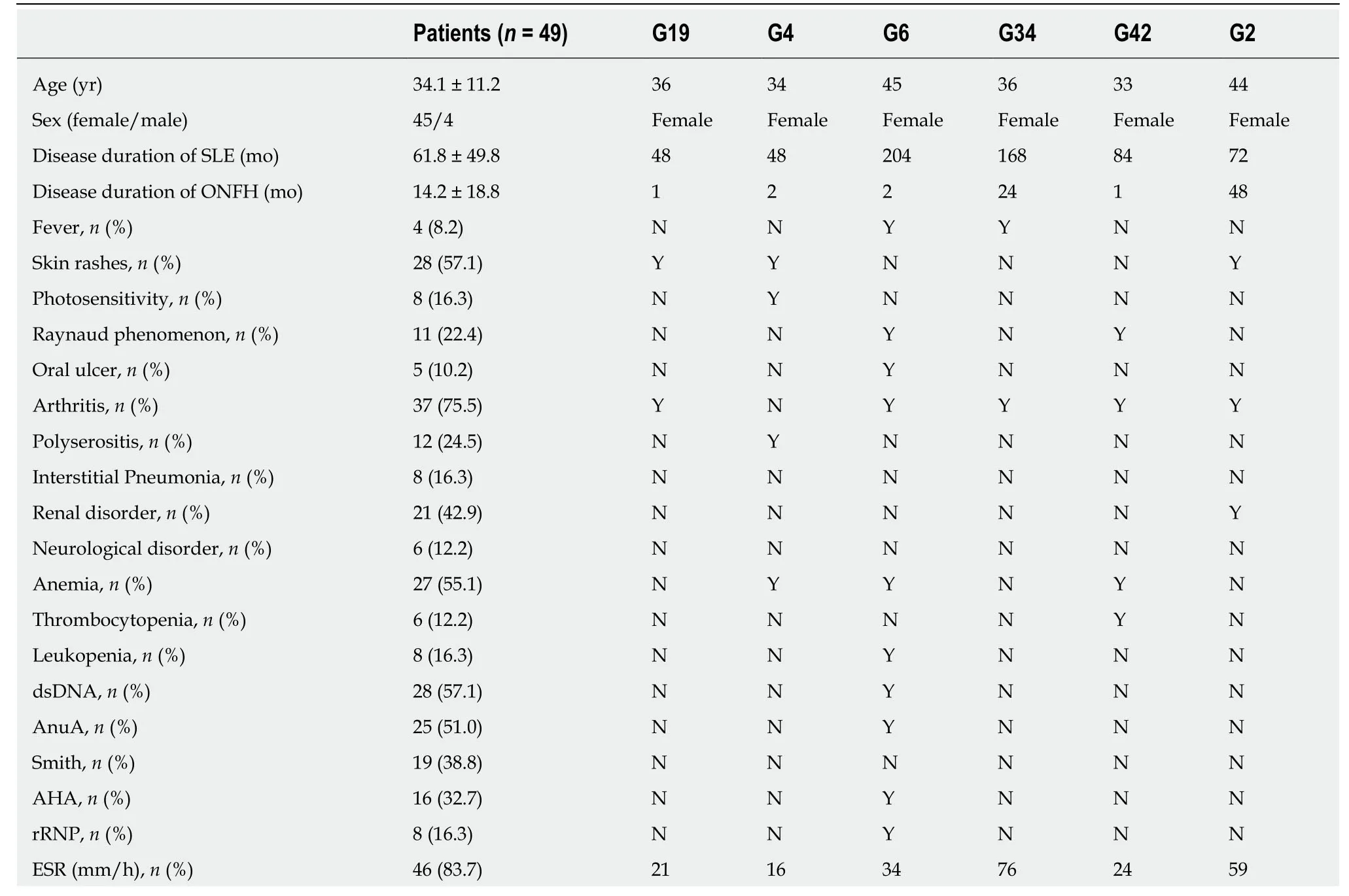
Table4 Demographics of patients with systemic lupus erythematosus with osteonecrosis of the femoral head
AHA:Antihistone antibody;AnuA:Antinucleosome antibody;C3:Complement 3;C4:Complement 4;dsDNA:Anti-double-stranded DNA antibody;ESR:Erythrocyte sedimentation rate;rRNP:Antiribosome ribonucleoprotein antibody;SLE:Systemic lupus erythematosus;SLEDAI:Systemic lupus erythematosus disease activity index;Smith:Anti-Smith antibody;Y:Yes or positive;N:No or negative.Except where otherwise indicated,values are expressed as the mean ± standard deviation.

Figure1 Map showing the nitric oxide synthase N-terminal domain with nitric oxide synthase 3 mutations identified in patients with osteonecrosis of the femoral head in systemic lupus erythematosus.Top:Diagram of the nitric oxide synthase 3 (NOS3) protein structure.Nitric oxide synthase (NOS) comprises a NOS N-terminal domain (amino acids 121-481),a flavodoxin/NOS domain (amino acids 522-698),a flavin adenine dinucleotide-binding,type 1 domain (amino acids 752-979),and an oxidoreductase flavin adenine dinucleotide/nicotinamide adenine dinucleotide (P)-binding domain (amino acids 1011-1123).Middle:The novel mutation of the NOS3 gene coding sequence identified in this study,located in exon 6 (encoding amino acids 675-816).Bottom:Mutated nucleotides in exon 6 of NOS3 are shown in orange.Mutated amino acids in the NOS N-terminal domain of NOS3 are shown in orange.FAD-binding:Flavin adenine dinucleotide-binding;NAD:Nicotinamide adenine dinucleotide.

Figure2 Map showing the collagen triple helix repeat domain with collagen type II alpha 1 chain mutations identified in patients with osteonecrosis of the femoral head in systemic lupus erythematosus.Top:Diagram of the collagen type II alpha 1 chain (COL2A1) protein structure.The COL2A1 comprises a von Willebrand Factor C (VWFC) domain (amino acids 34-89),a collagen triple helix repeat domain (amino acids 120-1219),and a fibrillar collagen C-terminal domain(amino acids 1254-1486).Middle:The novel mutations identified in the COL2A1 gene coding sequence in this study are located in exon 8 (encoding amino acids 531-609) and exon 29 (encoding amino acids 1888-1936).Bottom:Mutated nucleotides in exons 8 and 29 of the COL2A1 gene are shown in orange.Mutated amino acids in the VWFC and the collagen triple helix repeat domains of COL2A1 are shown in orange.
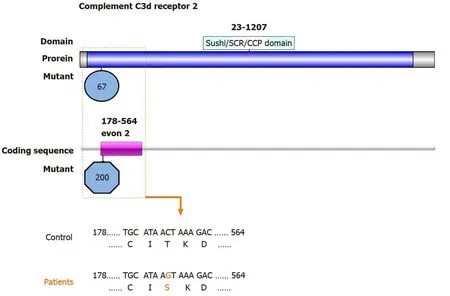
Figure3 Map of the Sushi/short consensus repeat/complement control protein domain of complement C3d receptor 2 with mutations identified in patients with osteonecrosis of the femoral head in systemic lupus erythematosus.Top:Diagram of the complement C3d receptor 2 (CR2) protein structure.CR2 comprises a Sushi/short consensus repeat/complement control protein domain (amino acids 23-1027).Middle:The novel mutation identified in the CR2 gene coding sequence in this study is located in exon 2 (encoding amino acids 178-564) Bottom:Mutated nucleotides in exon 2 of CR2 are shown in orange.Mutated amino acids in the Sushi/short consensus repeat/complement control protein domain of CR2 are shown in orange.CCP:Complement control protein;SCR:Short consensus repeat.
ARTICLE HIGHLIGHTS
Research background
Previous publications indicated that genetic predisposition might play important roles in the onset of osteonecrosis of the femoral head (ONFH) in systemic lupus erythematosus (SLE).
Research motivation
Complement C3d receptor 2 (CR2),nitric oxide synthase 3 (NOS3),collagen type II alpha 1 chain(COL2A1),protein tyrosine phosphatase non-receptor type 22 (PTPN22),and transient receptor potential cation channel subfamily V member 4 (TRPV4) were reported to be involved in the onset of ONFH in SLE.
Research objectives
To investigate whether the risk of ONFH in SLE is associated with single nucleotide variations(SNVs) inCR2,NOS3,COL2A1,PTPN22,andTRPV4.
Research methods
SNVs in theCR2,NOS3,COL2A1,PTPN22,andTRPV4genes were examined by using FastTarget and Illumina Miseq sequencing technologies in 49 cases of SLE with ONFH.Burrows-wheeler aligner was used to align the sequencing reads to hg19,and GATK and Varscan programs were used to perform SNV calling.PolyPhen-2,SIFT,and MutationTaster were used to assess the functional effects of non-synonymous SNVs.
Research results
Six patients were confirmed to have low frequency SNVs,including one patient with SNVs inNOS3(exon 6:c.814G>A:p.E272K and exon 7:c.814G>A:p.E272K.),four inCOL2A1(rs41263847:exon 29:c.1913C>T:p.T638I,exon 28:c.1706C>T:p.T569I,and rs371445823:exon 8:c.580G>A:p.A194T,exon 7:c.373G>A:p.A125T),and one inCR2(rs45573035:exon 2:c.200C>G:p.T67S).
Research conclusions
The onset of ONFH in SLE might be associated with the identified SNVs inNOS3,COL2A1,andCR2.And the low frequency functional mutations inNOS3had never been reported previously.
Research perspectives
These findings may have important pharmacogenetic implications.But the detailed mechanisms of the associations need to be determined in further studies.
 World Journal of Clinical Cases2020年12期
World Journal of Clinical Cases2020年12期
- World Journal of Clinical Cases的其它文章
- Assessment of diaphragmatic function by ultrasonography:Current approach and perspectives
- Computer navigation-assisted minimally invasive percutaneous screw placement for pelvic fractures
- Research on diagnosis-related group grouping of inpatient medical expenditure in colorectal cancer patients based on a decision tree model
- Evaluation of internal and shell stiffness in the differential diagnosis of breast non-mass lesions by shear wave elastography
- Real-time three-dimensional echocardiography predicts cardiotoxicity induced by postoperative chemotherapy in breast cancer patients
- Lenvatinib for large hepatocellular carcinomas with portal trunk invasion:Two case reports
