Effects of Euphorbiasteroid on gene expression in lung cancer cells based on gene chip
Zi-Ye Yang,Ming-Rui Jiang,Xiao-Tong Wei,Zhi-Cheng Wang,Zhu-Zhu Yue,Jing-Qiu Zhang,Meng-Yu Chen,Hui-Nan Wang,Meng-Lin Wang,Shuang-Hui Shi,Ying-Zi Wang*
1 School of Chinese Materia Medica,Beijing University of Chinese Medicine,102488,Beijing,China.
Abstract Objective Using gene chip technology to explore the molecular mechanism of euphorbiasteroid in the treatment of non-small cell lung cancer (NSCLC).Methods A549 cells were used as the in vitro model,and they were randomly divided into control group and different concentrations of euphorbiasteroid administration groups.Each group had 3 duplicate wells,after the cells were cultured in vitro,the cell viability was evaluated by CCK-8 method.Gene chip technology was used to screen the differentially expressed genes (DEGs)between the control group and the euphorbiasteroid administration group.The differential genes were further analyzed for Gene Ontology (GO) function enrichment and Kyoto Encyclopedia of Genes and Genome (KEGG) pathway enrichment analysis.The STRING online analysis platform combined with Cytoscape software to construct a target protein interaction (PPI) network and perform topological analysis to screen key targets,and use Real-time PCR (RTPCR) and molecular docking technology to verify key targets.Results According to the analysis of gene chip data,276 differentially expressed genes were screened,including 117 up-regulated genes and 159 down-regulated genes.GO analysis showed that differentially expressed genes were mainly involved in cell division,cell proliferation,cell cycle and other processes,involving protein binding,protein kinase binding and other functions,and were mainly distributed in nucleoplasm,chromosomes and other parts.KEGG signaling pathway analysis showed that differentially expressed genes were involved in cell cycle,pyrimidine metabolism,p53 signaling pathway and other pathways.PPI network analysis showed that CCNA2,TOP2A,CCNB1,CDC20,and RRM2 may be the key targets of euphorbiasteroid in the treatment of NSCLC.RT-PCR results showed that the expressions of CCNA2,TOP2A,CCNB1,CDC20,and RRM2 were significantly down-regulated in the euphorbiasteroid administered group,which was consistent with the gene chip results.Molecular docking results showed that euphorbiasteroid had good affinity with key targets and could bind spontaneously and stably.Conclusion The combination of gene chip,RT-PCR technology and molecular docking technology can find out the differential genes after the intervention of euphorbiasteroid,which can be used to explore the mechanism of euphorbiasteroid in the treatment of NSCLC.
Keywords Euphorbiasteroid;non-Small cell lung cancer;Gene chip;Differentially expressed genes;Molecular docking
Background
Lung cancer is one of the common malignant tumors,its mortality rate ranks first among all cancers.According to the clinical attributes of lung cancer,it is divided into two types: non-small cell lung cancer and small cell lung cancer,of which NSCLC accounts for more than 80% [1].At present,modern medical treatment of NSCLC mainly adopts chemotherapy,radiotherapy,immunotherapy and targeted therapy,but it is prone to recurrence and drug resistance [2,3].In recent years,with the deepening of the research on the treatment of malignant tumors by traditional Chinese medicine,traditional Chinese medicine has shown unique advantages in the treatment of NSCLC,which makes the clinical treatment of malignant tumors have a new choice.Chinese medicine Semen Euphorbiae is the dried ripe seeds of Euphorbiaceae plantEuphorbia lathyrisL.More than 30 kinds of diterpenoids have been isolated from Semen Euphorbiae,most of which are diterpene esters,among which euphorbiasteroid,also known as Euphorbia factor L1(EFL1),is the index component of Semen Euphorbiae in the 2020 edition of the Chinese Pharmacopoeia.Euphorbiasteroid has significant anti-tumor activity.It has been confirmed that it has certain toxicity to human lung cancer A549 cells.At the same time,it has a certain tumor inhibitory effect on Lewis non-small cell lung cancer model mice.It has the advantages of less damage to the body and less side effects [4-6].However,the molecular regulation mechanism of euphorbiasteroid in the treatment of non-small cell lung cancer has not yet been fully elucidated.
The development of gene chip technology has provided convenience for the study of multiple targets of traditional Chinese medicine,which can obtain a large number of differentially expressed genes in a short period of time.It is of great significance for molecular targeted therapy of cancer and interpretation of the mechanism of action of traditional Chinese medicine to mine the hub genes related to the treatment of tumors by traditional Chinese medicine through bioinformatics analysis.In this study,the gene chip technology was used to determine the gene expression profile of A549 cells intervened by euphorbiasteroid,and the differentially expressed genes were screened.The biological functions and pathways involved in the potential targets of euphorbiasteroid were speculated by GO and KEGG analysis,and verified by RT-PCR and molecular docking technology.The molecular mechanism and key targets of euphorbiasteroid for the treatment of non-small cell lung cancer were initially explored from the perspective of differential genes,which provided the basis for the clinical application of euphorbiasteroid.
Materials and methods
Experimental cells
Human lung cancer cell line A549 was purchased from ATCC cell bank.
Experimental drugs
Euphorbiasteroid,Lot: DSTDQ007001,Durst Biotechnology Co.,Ltd.,purity: 99.91%.Accurately weigh of euphorbiasteroid and add an appropriate amount of DMSO solution to prepare a mother solution with a mass concentration of 2.0 mg/mL,which is diluted in equal proportions with DMEM to obtain mass concentrations of 2×103,1×103,500,250,125,62.5,31.25,15.625,7.8125,3.90625,1.95313 μg/mL of the test solution,after each test is filtered with a 0.22 μm filter,for later use.
Experimental reagents
Cell proliferation detection kit (Shanghai Biyuntian Biotechnology Co.,Ltd.,Lot: C35006);DMEM medium(Thermo,USA,Lot: 10569044);fetal bovine serum(Thermo,USA,Lot: 10091-418);pancreatin (Thermo,USA,Lot: 15050057);penicillin-streptomycin double antibody (Thermo,USA,Lot: 15140122);PBS (Thermo,USA,Lot: C10010500BT);Trizol (Tiangen Biochemical Technology (Beijing) Co.,Ltd.,Lot: DP424);Reverse Transcription Kit (Thermo,USA,Lot: 00322404);PCR Kit (TaKaRa,Japan,Lot: AL61909A);GeneChip®Hybridization,Washing and Staining Kit (Affymetrix,USA,Lot: P/N900720);PCR primers were purchased from Invitrogen company of the United States;experimental water was prepared from Millipore water purifier.
Experimental equipment
Nano Drop ND-2000 UV spectrophotometer was purchased from Thermo (MA,USA).DYY-6C electrophoresis instrument was purchased from Beijing Liuyi Instrument Factory (Beijing,China).ABI7500 fluorescence quantitative PCR instrument,311 CO2constant temperature incubator and B004 microplate reader were purchased from Thermo (MA,USA).TS2 inverted phase contrast microscope was purchased from Nikon (Tokyo,Japan).HR1500-IIA2 biological safety cabinet was purchased from Haier (Qiandao,China).5424 centrifuge and Research micropipette were purchased from Eppendorf (Hamburg,Germany).BSI10S analytical balance was purchased from Beijing Sartorius Instrument Co.,Ltd.(Beijing,China);BD-C6 flow cytometer was purchased from Becton Dickinson and Company (New Jersey,USA).QL-902 vortex shaker was purchased from Haimen Qilin Bell Instrument Manufacturing Co.,Ltd.(Jiangsu,China).Tanon 1600 gel imaging system was purchased from Shanghai Tianneng Technology Co.,Ltd.(Shanghai,China).GeneChip Fluidics Station450,GeneChip Scanner 30007G and GeneChip Hybridization Oven645 were purchased from Affymetrix (California,USA).
Cell culture
Lung cancer A549 cells were routinely cultured in DMEM medium supplemented with 10% fetal bovine serum and 1%double antibody,and cultured in 37℃,5% CO2incubator.When the cell density reached 80%-90%,the medium was aspirated with a pipette,and the cells were rinsed 2-3 times with PBS buffer.Add pancreatin and place it in 37℃
incubators to digest for 1 minute.When most of the cells become round and fall off,quickly add a small amount of medium to terminate the digestion,and gently pipette evenly.Cell passage is carried out at 1:4,and the cells in good condition are taken for experiments.
CCK-8 assay to detect the survival rate of lung cancer cells
A549 cells in logarithmic growth phase were taken and seeded in a 96-well plate at a density of 5 × 103cells/well,and cultured overnight.When the cells were grown to confluence,they were divided into the euphorbiasteroid administration group and the control group.The administration groups were respectively added with the test solution of 2000,1000,500,250,125,62.5,31.25,15.625,7.8125,3.90625,1.95313 μg/mL of the test solution of euphorbiasteroid;the control group was added with the same volume of DMEM for culture base.Three duplicate wells were set in each group,and 10 μL of CCK8 solution was added to each well after 48 h of treatment.Incubate at 37℃ for 1 h,using a microplate reader to measure the absorbance at 450 nm,and calculate the cell viability according to the formula:
Total RNA extraction
The total RNA of lung cancer A549 cells was extracted by the Trizol method,the medium in the 96-well plate was aspirated,and 1 mL of 4℃ pre-cooled PBS solution was added to each well for washing twice,the PBS was discarded,and 1 mL of Trizol lysis solution was added to each well,repeat the pipetting until the cells are completely lysed.Collect the cell lysate in a 1.5 mL RNase-free sterile centrifuge tube and let it stand on ice;add 0.2 mL of chloroform to a 1.5 mL centrifuge tube,place the centrifuge tube on a micro-vortex shaker,and shake until it appears emulsified and stand still.Centrifuge the tube at 4℃ and 12000 g for 15 min.Take the supernatant and put it in a 1.5 mL RNase-free sterile centrifuge tube,add 0.5 mL of isopropanol into the centrifuge tube,mix by pipetting repeatedly,incubate at room temperature for 10 min,and place the centrifuge tube at 4℃ and 12000 g for 10 min.Discard the supernatant,75% ethanol solution was added to wash the precipitate,and the centrifuge tube was centrifuged at 4℃ and 12000 g for 10 min.Discard the supernatant and add an appropriate amount of RNase-free H2O to dissolve the precipitate.
Sample detection
The total RNA extracted above was DNA digested with DNaseI solution,and then the samples were analyzed for DNA contamination by agarose electrophoresis.Then use UV spectrophotometer and Agilent 2100 Bioanalyzer to determine the integrity of RNA,and its integrity judgment standard is A260/280 ≥ 1.80;28S: 18S rRNA band brightness greater than 0.7.After passing the above inspection,the follow-up gene chip test can be carried out.
The preparation of gene chip
Starting with the total RNA of the sample,the detection of the gene chip is performed,and the total RNA is amplified in vitro and labeled with biotin.The cRNA amplification labeling kit is used for the labeling process of biotin.
(1) Reverse transcription synthesis of First strand cDNA:Starting with total RNA,containing T7Oligo(dT) Primer as primers,first strand cDNA was synthesized using the first strand enzyme mixture.
(2) Synthesis of Second strand cDNA: Use the second strand enzyme mixture to convert the RNA strand in the DNA-RNA into Second strand cDNA to synthesize double-stranded DNA.
(3) in vitro transcription to synthesize cRNA: Using Second strand cDNA as a template,and T7 EnzymeMix to synthesize cRNA and incorporate biotin.
(4) cRNA purification: Use magnetic beads to purify the cRNA,remove impurities such as salts,enzymes,and quantify cRNA.
(5) cRNA fragmentation: Fragment the cRNA to a suitable size for hybridization
(6) Fragmented biotin-labeled cRNA and chips were hybridized in a GeneChip Hybridization Oven 640 hybridization oven (Affymetrix) at 45℃ for 16 h with rotation.
(7) The hybridized chip was washed and stained on the GeneChip Fluidics Station 450 workstation (Affymetrix).The chip was then scanned with a GeneChip Scanner3000 scanner (Affymetrix) to obtain a chip hybridization map.
Data extraction and statistical analysis
AGCC software (Affymetrix®GeneChip®Command Console®Software) was used to analyze the chip image,so that the image signal,namely.DAT file was converted into a.CEL file,that is,digital signal,and the.CEL file recorded the fluorescence signal intensity at the probe level.The.CEL file is used as the source file for data preprocessing,and the RMA algorithm is used for data preprocessing.
Differential gene and functional analysis
Differential genes are the main objects of gene expression profiling,which usually refer to genes that express significantly different in different tissues,and it is also an important manifestation of cell changes after euphorbiasteroid administration.The differentially expressed genes were analyzed on the preprocessed data,and the differential genes were screened usingP-value and Fold Change (FC) values.The screening criteria for differential genes were:P-value ≤ 0.05 and |log2FC| > 2,the differential genes that were significantly up-regulated and down-regulated in the administration group after treatment with euphorbiasteroid compared with the control group were obtained,and the differentially expressed genes were visually represented by volcano plots overall distribution.Multiple groups of differential genes are generally clustered analysis in the form of heat maps,which can study the changes in the expression of differential genes in multiple groups of samples.
GO is a database established by the Gene Ontology Consortium,which is mainly used to define and describe the functions of genes and proteins.GO functional analysis includes: Biological Process,Cellular Component and Molecular Function,which describe the biological processes involved in genes and their products,the cellular environment in which they are located,and the molecular functions they can perform,etc.In this experiment,GO function enrichment analysis was performed on the differentially expressed genes of the administration group and the control group,and the GO number corresponding to each gene was found by the method of ID correspondence or sequence alignment.Then,the number corresponds to each entry in the GO database,and the number of genes on each entry is calculated to obtain GO entries with significantly enriched differentially expressed genes.At the same time,the R software cluster Profiler was used for GO enrichment analysis and data visualization.
KEGG is a database that analyzes the metabolic pathways and functions of gene products in cells.Pathway enrichment analysis is performed on differential genes to understand the process of genes performing biological functions.After setting cutoff screening of differential analysis to obtain significantly differential genes,enrichment analysis is performed based on hypergeometric distribution,and compared with the background of the entire genome,the Pathways with significantly enriched differential genes are screened out.P-value ≤ 0.05 was defined as a KEGG pathway with significant enrichment of differentially expressed genes.Pathway enrichment analysis and data visualization were performed using the R software cluster Profiler.
PPI network construction and key target screening
The STRING database (www.string-db.org) collects and integrates known and predicted protein-protein association data for many organisms,including humans.The screened differential genes were imported into the STRING database,and PPI were constructed under the condition that the combination score > 0.4.The TSV data files were downloaded and imported into Cytoscape software for visualization,and the Cytohubba plugin was used for topological analysis to screen key hub genes.The Cytohubba plugin provides features including edge percolated component (EPC),Degree,maximum neighborhood component (MNC),density of maximum neighborhood component (DMNC) and maximum clique centrality (MCC) and other topological analysis methods.In order to improve the accuracy of the prediction results,this study explored the top 10 important node genes based on EPC,Degree and MNC,respectively,and took the intersection,obtained the key genes for the treatment of lung cancer with euphorbiasteroid.
RT-PCR detection of key differential gene expression
Collect the A549 cells from the control group and the administration group respectively.According to the procedures in "Total RNA extraction" extract total RNA,and use NanoDrop® ND-2000 to determine RNA concentration and purity.Then use PrimeScript™ RT reagent Kit with gDNA Eraser for cDNA reverse transcription and RT-PCR for amplification.
The reaction conditions are: 95℃ for 30s,1 cycle;95℃for 5s,60℃ for 40s,a total of 40 PCR cycles.In order to establish the melting curve of the PCR product,after the amplification reaction,press 95℃ for 10s;60℃ for 60s;95℃ for 15s;and slowly heat from 60℃ to 99℃.Taking GAPDH as an internal reference,its relative expression was calculated by the 2-ΔΔCtmethod.The PCR primer sequences are shown in Table 1.
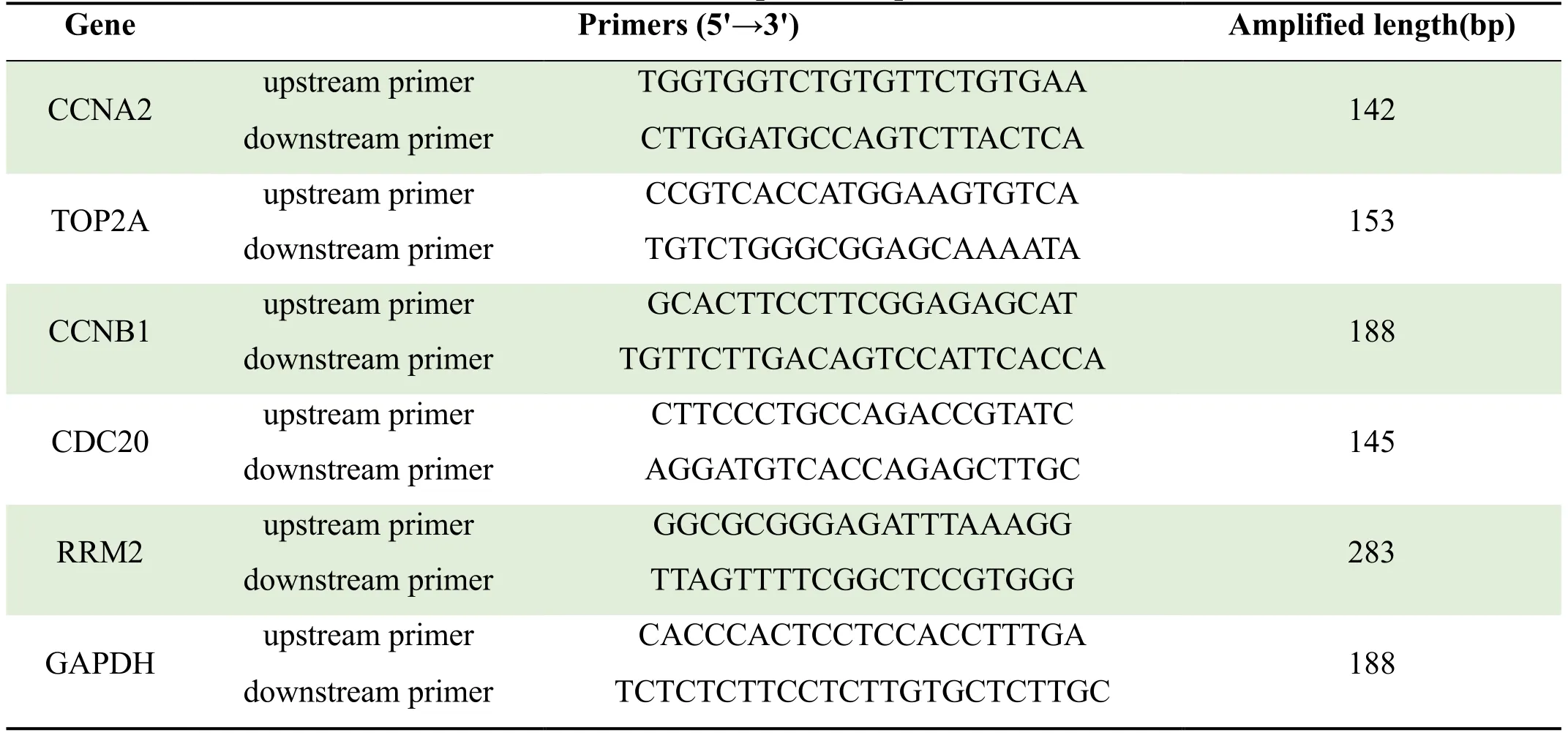
Table 1 PCR primer sequences
Molecular docking verification
Molecular docking verification was performed on the screened key genes using Auto dock vina (v1.1.2).Download the SDF format of the 3D structure of euphorbiasteroid in the PubChem database and convert it to MOL2 format,and download the three-dimensional protein structure of key genes from the PDB protein database.Preprocessing of compounds and proteins using PyMol (v1.7.2.1) and Auto dock tools (v1.5.6),including identified compound rotatable bonds,removed protein water molecules,other ligand small molecules,hydrogenation,incorporation of nonpolar hydrogen,etc.,and save compound and protein files in pdbqt format.Use the plugin Grid Box in Auto dock tools to set the docking area,perform molecular docking,and use PyMol for visualization.
Statistical methods
The experimental results were statistically analyzed using SAS 9.4 and GraphPad Prism 8.0.If each group of data had a normal distribution and homogeneity of variance,quantitative data were presented as the means ± standard deviations.One-way analysis of variance was used for comparison between multiple groups,LSD t-test was used for pairwise comparison within groups,and one-way analysis of variance (ANOVA) was used for comparison between multiple groups;P<0.05 was considered statistically significant.
Results
Effect of Euphobiasteroid on proliferation of lung cancer cells
CCK-8 assay result showed that the survival rate of A459 cells was decreased in a dose-dependent manner in the concentration range of 0 ~ 2.00 mg/mL (P<0.05)compared with that of the control group after 48 h treatment with each test substance of Euphobiasteroid .The results are shown in Table 1 and Figure 1.In addition,the IC50of Euphobiasteroid on A549 cells was 24.94 μg/mL,so 24.94 μg/mL concentration of Euphobiasteroid was selected for subsequent gene chip sequencing and RTPCR experiments.
Table 2 The effect of Euphobiasteroid on the survival rate of A549 cells ( ± s,n=5 )
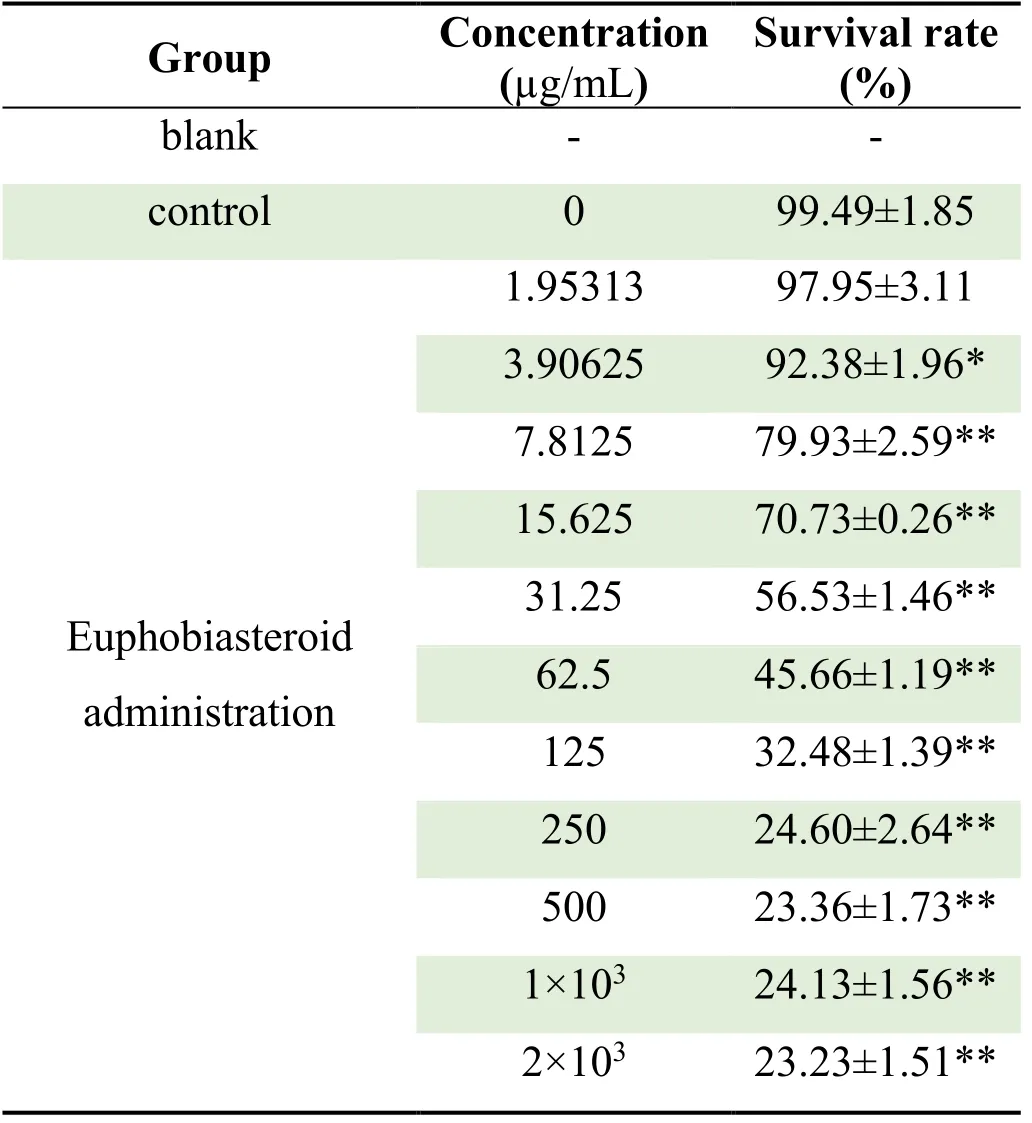
Table 2 The effect of Euphobiasteroid on the survival rate of A549 cells ( ± s,n=5 )
Note: Compared with the control group,*P<0.05,**P<0.01
Group Concentration(μg/mL)Survival rate(%)blank - -control 0 99.49±1.85 1.95313 97.95±3.11 3.90625 92.38±1.96*7.8125 79.93±2.59**15.625 70.73±0.26**Euphobiasteroid administration 31.25 56.53±1.46**62.5 45.66±1.19**125 32.48±1.39**250 24.60±2.64**500 23.36±1.73**1×103 24.13±1.56**2×103 23.23±1.51**
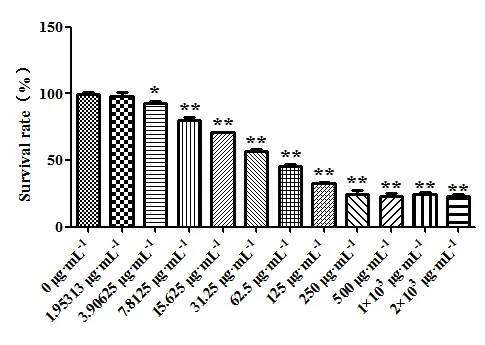
Figure 1 Effects of different concentrations of Euphobiasteroid on proliferation of A549 cells.(n=5;Compared with the control group,*P<0.05,**P<0.01)
Effect of Euphobiasteroid on lung cancer cell gene chip RNA quality test results
The total RNA extracted from each group was detected by Nanodrop2000 spectrophotometer and Agilent2100 Bioanalyzer,and A260/280 was between 1.93 and 2.01,indicating that there was no protein residue in RNA samples.The 28S rRNA and 18S rRNA bands of RNA sample electrophoresis was clearly visible by formaldehyde denaturing agarose gel electrophoresis,indicating that RNA was not degraded and had good integrity.The quality met the requirements of the microarray experiment and could be used for subsequent experiments.The results are shown in Table 3 and Figure 2.
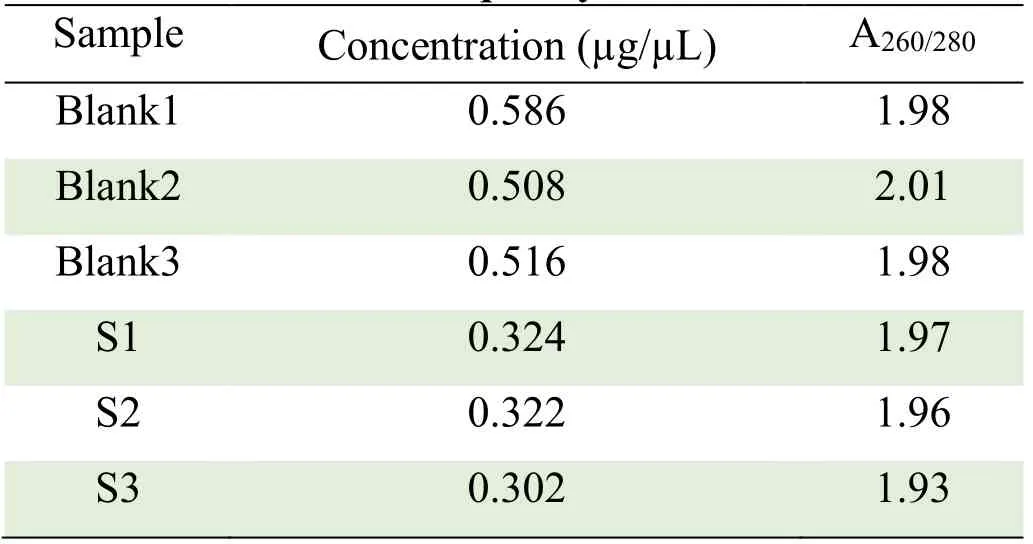
Table 3 RNA quality test results
Differentially expressed genes
WithP<0.05,|log2FC| > 2 as the standard,the differentially expressed genes (DEGs) were screened as the target genes for the treatment of non-small cell lung cancer with Euphobiasteroid .A total of 276 DEGs were screened out in the control group and Euphobiasteroid administration group,of which 117 were significantly upregulated and 159 were significantly down-regulated.R software (version 4.1.0) was used to draw heat map and volcano map using the Plot function and Pheatmap package respectively,clustered heat map can study the expression of DEGs in the control group and the Euphobiasteroid administration group samples (Figure 3).DEGs were shown in volcanic map (Figure 4).Red dots represent up-regulated genes,blue dots represent downregulated genes,and gray dots represent non-differentially expressed genes.
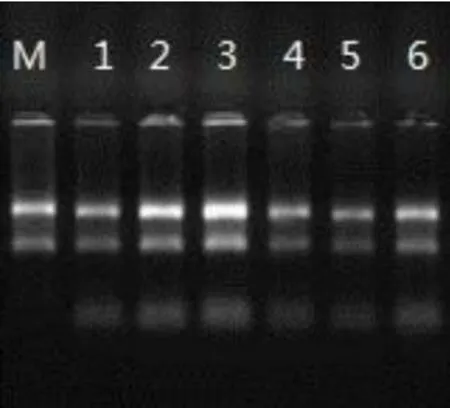
Figure 2 Sample quality electrophoresis.(1-3: blank group;4-5: Euphobiasteroid administration group)
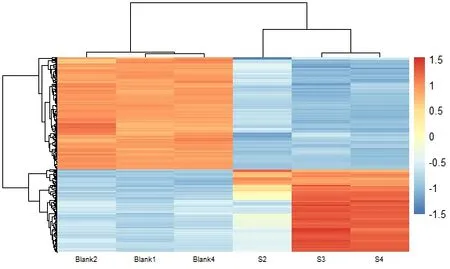
Figure 3 Clustering Heat Map of DEGs.(Blank1,2,4:control group;S2,3,4: Euphobiasteroid administration group)
Functional enrichment analysis of DEGs
In order to further explain the biological function of the DEGs after Euphobiasteroid intervention in lung cancer A549 cells,GO functional enrichment analysis was carried out for the screened DEGs,withP-Value > 0.05 as the index,a total of 157 entries were obtained by GO enrichment analysis,including 93 biological processes,22 molecular functions,and 42 cellular components.And according to thePvalue sorting,select the top 10 entries for each module,as shown in Figure 5-7.The results showed that the DEGs had an impact on biological process,cellular component and molecular function.
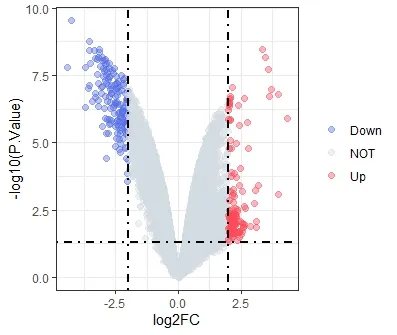
Figure 4 Volcano map.(Red is the up-regulated genes;blue is the down-regulated genes)
In the biological process,DEGs mainly involve cell division,DNA replication,G1/S transition of mitotic cell cycle,chromosome segregation,cell proliferation and cell cycle,etc.In the molecular function,it mainly affects protein binding,single-stranded DNA binding,ATP binding,protein kinase binding,and kinesin binding,etc.In the cellular component,DEGs are mainly located in nucleoplasm,chromosome,centromeric region,spindle microtubule,cytoplasm,chromatin,MCM complex,etc.
KEGG pathway analysis of DEGs
The KEGG database integrates the information of genome,chemistry and system functions,and can associate the DEGs with the system functions at the level of cells,species and even ecosystem,so as to understand the biological processes of genes in organisms through mutual cooperation.The enrichment analysis results of KEGG pathway were sorted according to thePvalue from small to large,and the pathway was plotted as bubbles,as shown in Figure 8.There were 8 pathways involved in 276 DEGs screened between the Euphobiasteroid treatment group and the control group.Including cell cycle,DNA replication,Oocyte meiosis,Pyrimidine metabolism,progesterone-mediated oocyte maturation,p53 signaling pathway,Nucleotide excision repair and one carbon pool by folate.
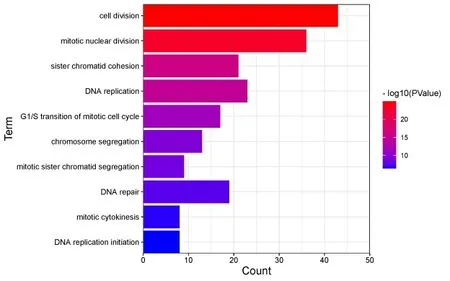
Figure 5 Biological Process enrichment diagram
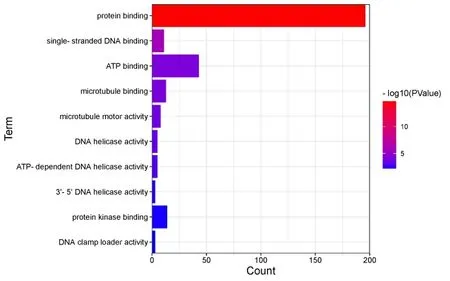
Figure 6 Molecular Function enrichment diagram
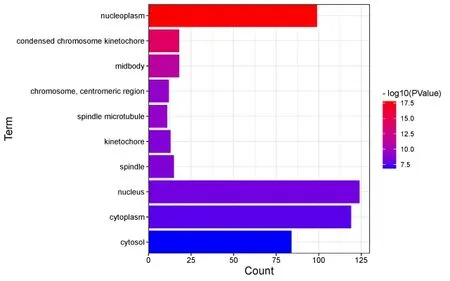
Figure 7 Cellular Component enrichment diagram
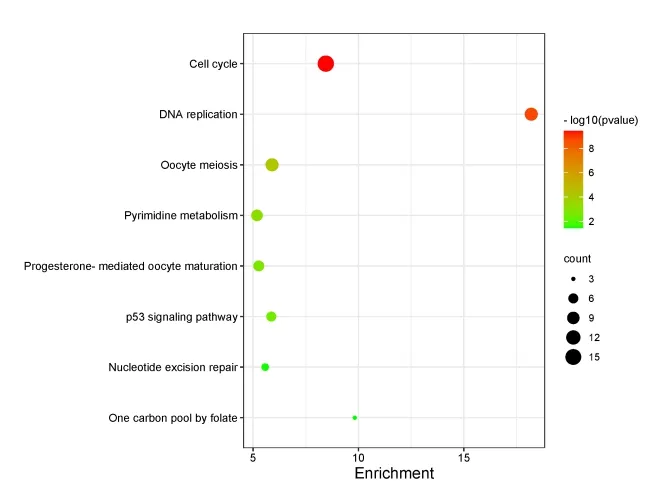
Figure 8 Enrichment analysis of KEGG pathway
Construction of PPI network and screening of key hub genes
Totally,276 DEGs were imported into STRING database to obtain the interaction network between differential gene-coded proteins and proteins.The TSV format of the results was imported into Cytoscape software to construct the interaction network between visual proteins,as shown in Figure 9.The PPI network has 276 nodes,4946 edges,the average clustering correlation coefficient is 0.596,PPI enrichment<1.0 e-16.The three algorithms like Edge Percolated Component (EPC),Degree and Maximum Neighborhood Component (MNC) in Cytoscape plug-in cytohubba were combined to screen the top 10 important nodes in PPI network,and Venn diagram was used to show the screening results,as shown in Figure 10.Finally,CCNA2,TOP2A,CCNB1,CDC20 and RRM2 were obtained,which were all significantly down-regulated,that is,the key genes for the treatment of non-small cell lung cancer with Euphobiasteroid .
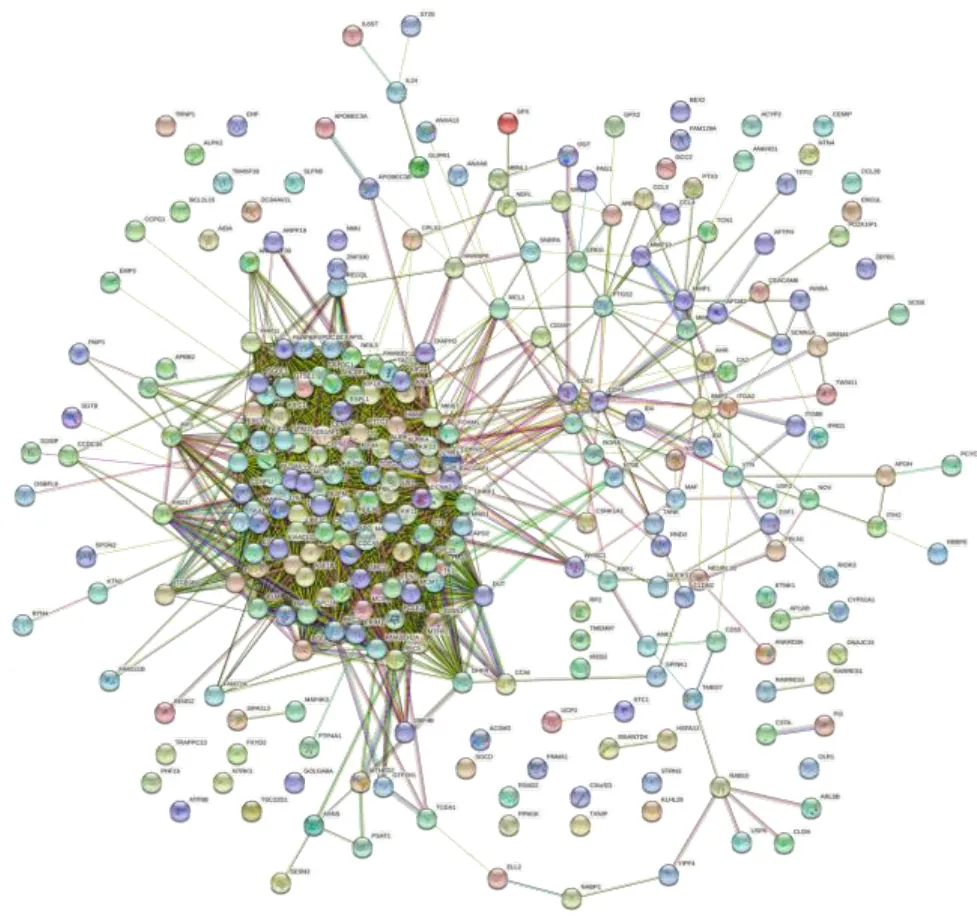
Figure 9 PPI network of DEGs
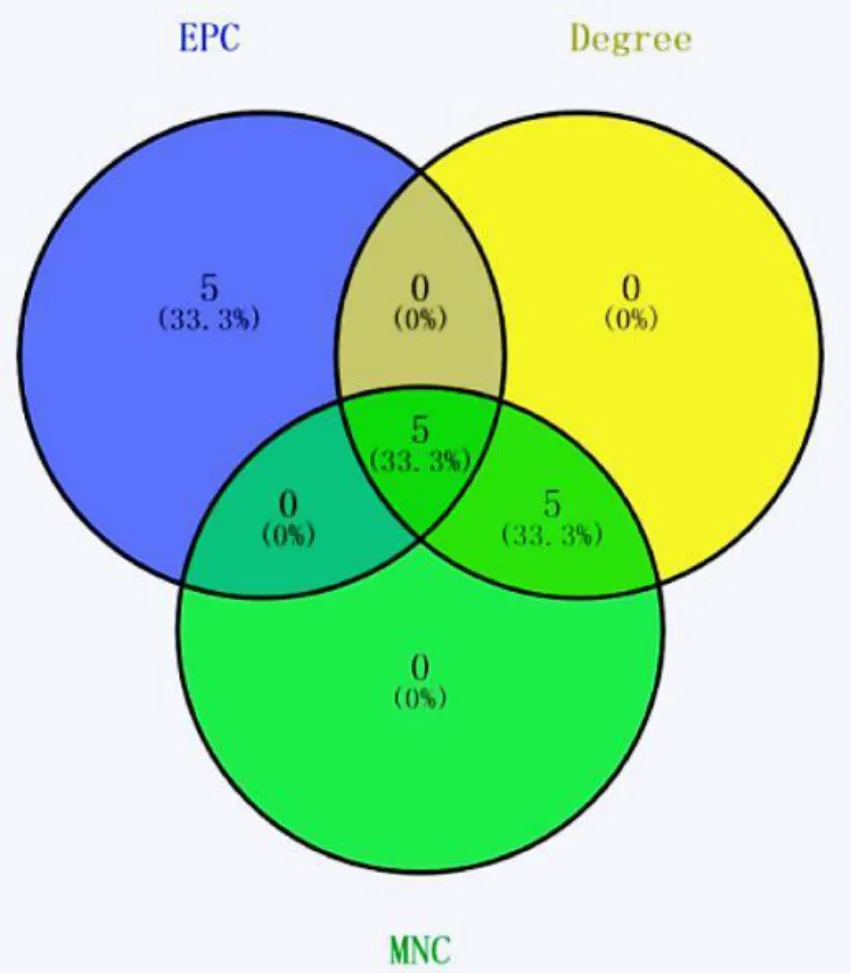
Figure 10 The Venn diagram of TOP10 gene in EPC,Degree and MNC algorithm
RT-PCR detection of transcription factor mRNA expression
In order to verify the accuracy of gene chip results,five key genes were selected for qRT-PCR detection.The mRNA expression levels of CCNA2,CCNB1,TOP2A,CDC20 and RRM2 in A549 cells of each group were detected by RT-PCR.The results showed that the mRNA expression levels of CCNA2,CCNB1,TOP2A,CDC20 and RRM2 in A549 cells of the Euphobiasteroid administration group were significantly decreased compared with the blank group (P<0.01),which was consistent with the gene chip results,indicating that the sequencing results were accurate and reliable,as shown in Figure 11.
Molecular docking verification results
In this study,five selected key genes CCNA2,CCNB1,TOP2A,CDC20 and RRM2 were selected as receptors for molecular docking verification.The results were shown in Table 4 and Figure 12.The binding energies were all<0,indicating that Euphobiasteroid and five target receptors can spontaneously bind,and the binding energies were all<-5,indicating that there is a strong binding activity.Thesmaller the value,the easier the Euphobiasteroid bind to the target.
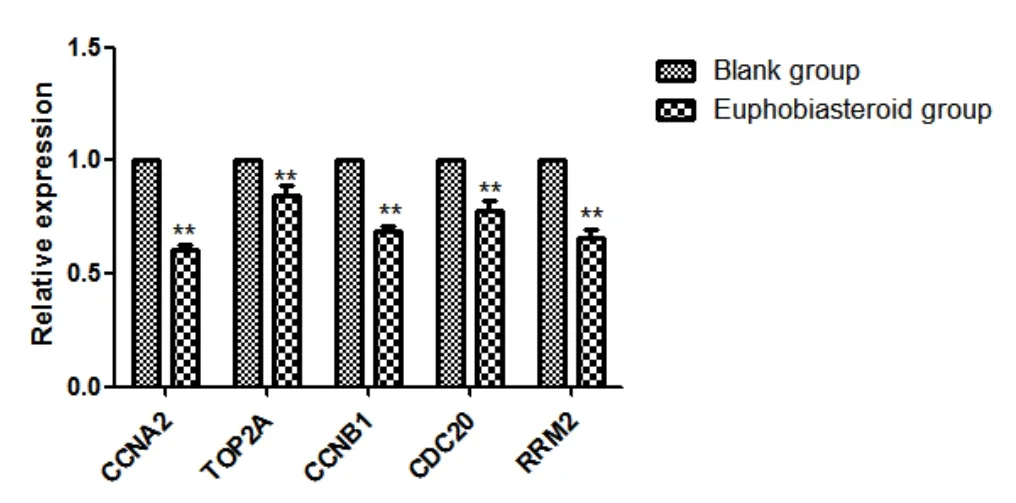
Figure 11 Effects of Euphobiasteroid on mRNA expression of CCNA2,CCNB1,TOP2A,CDC20 and RRM2 in A549 cells
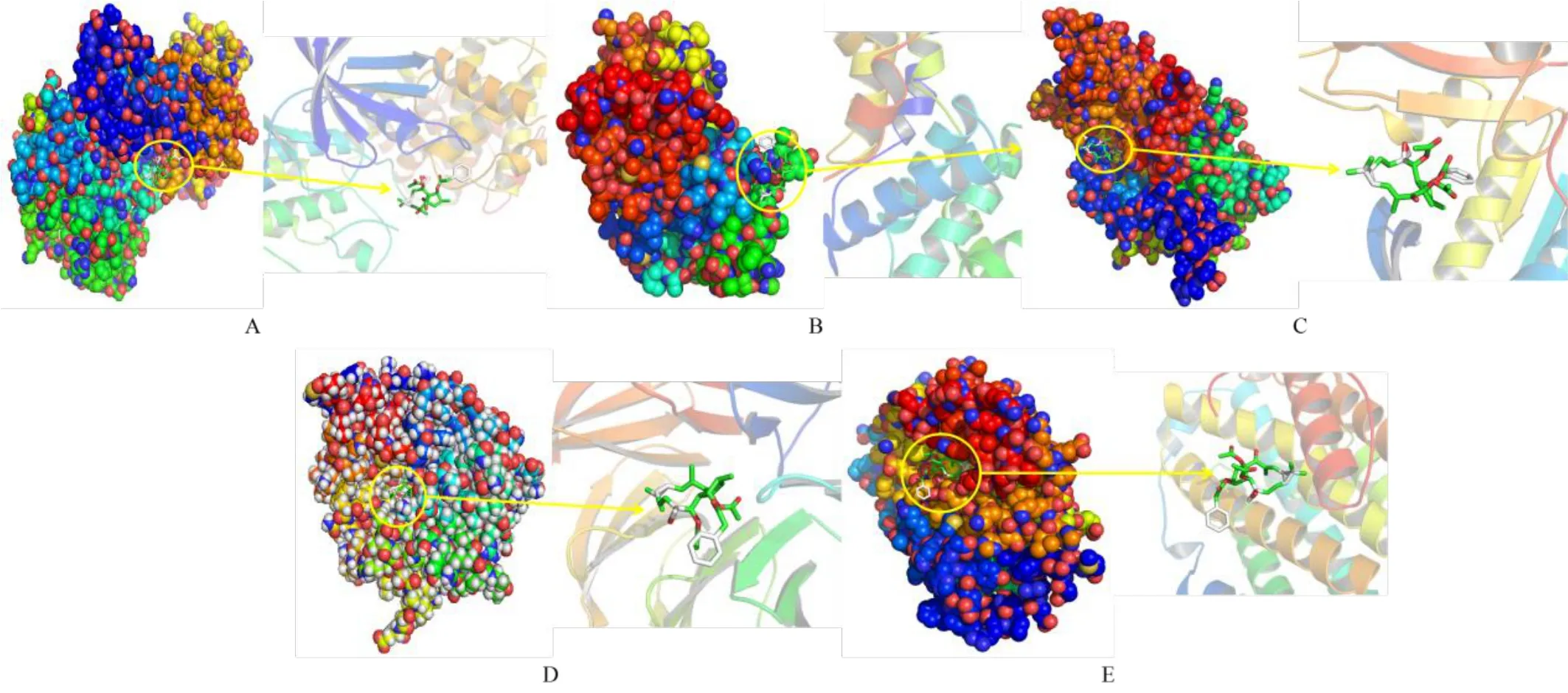
Figure 12 Visual molecular docking results of Euphobiasteroid binding to the key targets.(A.CCNA2;B.CCNB1;C.TOP2A;D.CDC20;E.RRM2)
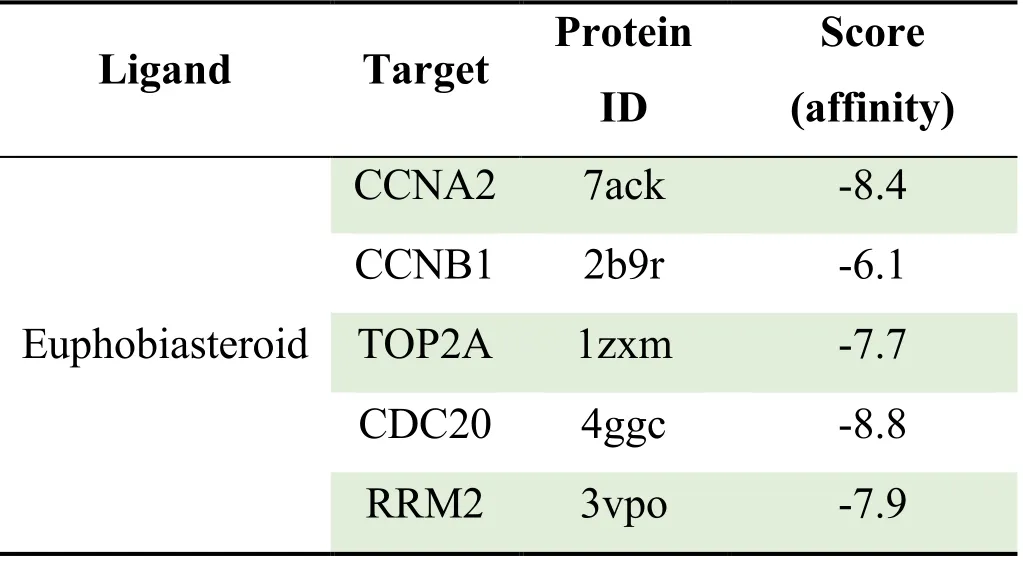
Table 4 The results of Euphobiasteroid and key targets molecular docking
Discussion
In this study,through the analysis of DEGs,it was found that compared with normal A549 cells,a total of 276 DEGs were obtained after Euphobiasteroid intervention,including 117 up-regulated genes and 159 down-regulated genes.The PPI network construction and topology analysis of 276 DEGs showed that CCNA2,TOP2 A,CCNB1,CDC20 and RRM2 had higher degree values,which might be the potential core targets for the treatment of NSCLC with Euphobiasteroid.Among them,CCNA2 and CCNB1 are members of the cyclin family [7-11],which play an important role in regulating cell growth,proliferation and apoptosis.Some studies have reported that CCNA2 and CCNB1 are closely related to the metastasis and malignant progression of non-small cell lung cancer,and are highly expressed in tumors.TOP2A is an enzyme that exists in cells [12-17].It regulates mitosis by catalyzing the cleavage of double-stranded DNA,and high expression of TOP2A is detected in non-small cell lung cancer,breast cancer and other cancers,which is considered as a cancer target in clinical application.CDC20 is a key gene regulating cell cycle and apoptosis [18-21].As an oncogene,CDC20 is overexpressed in a variety of human tumor tissues,and can promote the occurrence and development of tumors by promoting epithelialmesenchymal transition.RRM2 is highly expressed in a variety of tumor tissues by involving carcinoma associated kinases affecting RNA transport and oocyte meiosis.The high expression level of RRM2 is closely related to tumor cell migration and invasion,and has a certain impact on tumor metastasis and prognosis [22-25].In this study,compared with the control group,the mRNA expression levels of CCNA2,TOP2 A,CCNB1,CDC20 and RRM2 in the Euphobiasteroid treatment group were significantly decreased,and the molecular docking results showed that Euphobiasteroid had strong binding activity with these target proteins,indicating that CCNA2,TOP2 A,CCNB1,CDC20 and RRM2 targets could be used as potential targets for Euphobiasteroid in the treatment of NSCLC.
The occurrence of NSCLC is caused by many factors,and its progression is also affected by multiple paths.Multipathway analysis showed that DEGs were enriched in cell cycle pathway,pyrimidine metabolism pathway and p53 signaling pathway,etc.Among them,the regulation of cell cycle pathway is the main symbol of anti-cancer effect,and the cycle regulation of cancer cells is closely related to the occurrence and development of cancer.Pathogenic factors can induce the activation of oncogenes and the inactivation of tumor suppressor genes through various cell signal transduction,thereby regulating the cell cycle,promoting the proliferation of cancer cells and inhibiting the apoptosis of cancer cells [26].The pyrimidine metabolism pathway is the key pathway for the energy generation of cancer cells [27].The characteristics of cancer cells are the abnormal increase in energy production and biosynthesis to maintain their growth and proliferation[28].The pyrimidine metabolism belongs to nucleotide metabolism,which can produce pyrimidine molecules to provide the necessary components for DNA replication and RNA synthesis of cancer cells,and provide corresponding auxiliary factors and energy to promote the proliferation of cancer cells [29].In addition,some studies have shown that the content of pyrimidine in highly invasive cancer cells exceeds the content required for proliferation,and it is found that excessive pyrimidine may be degraded to promote epithelial-mesenchymal transition,thereby enhancing the invasion and differentiation of cancer cells [30-32].Therefore,Euphobiasteroid may affect the energy production and epithelial-mesenchymal transition of A549 lung cancer cells by interfering with the pyrimidine metabolic pathway,thereby inhibiting cell proliferation and reducing its invasion ability.As a classic tumor suppressor pathway,the role of p53 signaling pathway in various types of tumors has been widely studied.It is involved in regulating the proliferation,cycle,apoptosis and DNA damage of cancer cells [33,34],and is closely related to the occurrence and development of malignant tumors [35,36].This suggests that Euphobiasteroid may play a therapeutic role in NSCLC by inhibiting cell proliferation,invasion,migration,energy metabolism and promoting apoptosis.
Conclusions
In this study,276 DEGs and 5 key hub genes were screened by gene chip technology combined with bioinformatics methods.These genes may play an important role in the occurrence and development of NSCLC.The results can provide a theoretical basis for the clinical application of Euphobiasteroid in anti-tumor.
Data availability or Code availability
Correspondence and requests for materials should be addressed to wangyzi@sina.com (Ying-Zi Wang).
Abbreviations
NSCLC,non-small cell lung cancer;EFL1,Euphorbia factor L1;DEGs,differentially expressed genes;GO,Gene Ontology;KEGG,Kyoto Encyclopedia of Genes and Genomes;PPI,protein-protein interaction;RT PCR,Realtime PCR;FC,Fold Change;EPC,edge percolated component;MNC,maximum neighborhood component;DMNC,density of maximum neighborhood component;MCC,maximum clique centrality;ANOVA,one-way analysis of variance;CCNA2,Cyclin-A2;TOP2A,DNA topoisomerase 2-alpha;CCNB1,Cyclin-B1;CDC20,Cell division cycle protein 20 homolog;RRM2,Ribonucleoside-diphosphate reductase subunit M2.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (Grant No.2018YFE0197900).
Author contributions
Zi-Ye Yang,carried out the lab work,participated in the design of the study,and drafted the manuscript.Ming-Rui Jiang,Xiao-Tong Wei,Zhu-Zhu Yue,Zhi-Cheng Wang,Shuang-Hui Shi,and Meng-Lin Wang performed the experiments and carried out the statistical analyses.Hui-Nan Wang,Jing-Qiu Zhang,and Meng-Yu Chen collected the field data and critically revised the manuscript.Ying-Zi Wang conceived the study,designed the study,coordinated the study,and helped draft the manuscript.All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
 TMR Modern Herbal Medicine2022年3期
TMR Modern Herbal Medicine2022年3期
- TMR Modern Herbal Medicine的其它文章
- Traditional Chinese medicine as monotherapy or combined with oseltamivir in the treatment of H1N1 influenza: a systematic review and meta-analysis
- The genus Strobilanthes: phytochemistry and pharmacology
- Niaoduqing granules inhibits TGF-β1-induced epithelial-mesenchymal transition in human renal tubular epithelial HK-2 cells
- Standardization and Quality control parameters of Kayam Churna
- A review of the ethnobotanical value,phytochemistry,and pharmacology of Physalis pubescens L.
