Characterization of a novel DUF1618 gene family in rice
Lan Wang,Rongxin Shen,Le-Tian Chen,3* and Yao-Guang Liu*
1State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources,College of Life Sciences,South China Agricultural University,Guangzhou 510642,China,2College of Agriculture,South China Agricultural University,Guangzhou 510642,China,3Guangdong Provincial Key Laboratory of Protein Function and Regulation in Agricultural Organisms,College of Life Sciences,South China Agricultural University,Guangzhou 510642,China.*Correspondence:lotichen@scau.edu.cn;ygliu@scau.edu.cn
INTRODUCTION
Evolution of gene families is important for speciation and adaptation to environments(Ohta 2000);examination of these gene families can provide insights into mechanisms of development,adaptation,and evolution.For example,in the important cereal crop rice(Oryza sativa L.),sequencing and annotation of the genome have revealed a number of previously unknown gene families(Ohyanagi et al.2006),including a large set of gene families with domains of unknown function(DUF).Domains of unknown function protein families are named after their order of addition to the protein-family(Pfam)database since 1997,and the DUF numbering scheme now extends to DUF2607(Bateman et al.2010).With tremendous effort,some DUF genes have been experimentally investigated and renamed.For example,DUF1 and DUF2 have been found to act as enzymes to process the ubiquitous signaling molecule c-di-GMP and therefore have been designated as GGDEF and EAL,respectively(Romling and Simm 2009).DUF27,also called the macro domain,possesses an ADP-ribose binding activity(Karras et al.2005).DUF283 is a functional domain in Dicer or Dicer-like(DCL)proteins and plays essential roles in siRNA processing for gene silencing(Dlakic 2006;Qin et al.2010).Also,root UV-B sensitive 1(RUS1)encodes a DUF647 protein involved in UV-B sensing and early morphogenesis of seedlings(Tong et al.2008).Crystal structure analysis revealed an ABATE domain in a DUF1470 protein and suggested a role as a stress-induced transcriptional regulator(Bakolitsa et al.2010).
DUF1618 domain-containing genes belong to a large gene family containing a conserved domain of 56–199 amino acids(aa).Our analysis of present databases shows that this gene family is present only in several monocot plant species,suggesting it to be a relatively young gene family.However,this gene family has not been characterized,and no member has been functionally studied.In this study,we identified 121 DUF1618 members in the rice genome that are divided into two groups,with two clades in each group,based on phylogenetic analysis.Most DUF1618 genes in rice are expressed at very low levels.The experimental validation indicated that some DUF1618 genes have different expression patterns in different rice cultivars,and some members may be important regulators of biotic and abiotic stress responses.
RESULTS
Genome-wide survey and phylogenetic analysis of DUF1618 genes in rice
In order to extend our understanding of the DUF1618 gene family,we searched public databases through the National Center for Biotechnology Information(NCBI)website and found 468 rice protein sequence entries with a predicted DUF1618 domain.To eliminate redundancies,we analyzed these protein sequences and found that some of the protein entries are encoded by the same genes.We also confirmed the presence of a DUF1618 domain in each putative DUF1618 protein by using the Simple Modular Architecture Research Tool(SMART)in PFam domain analysis,and found that some of the protein entries did not contain a DUF1618 domain.Finally,a set of 121 non-redundant DUF1618 genes was identified in the rice genome(Figure 1).
Next,we sorted all these DUF1618 genes according to their location on chromosomes 1–12,and designated them with sequential numbers.To examine the phylogenetic relationship of these DUF1618 proteins,an unrooted phylogenetic tree was constructed from alignments of the DUF1618 domain sequences with MEGA 4.0(Tamura et al.2007).These DUF1618 proteins were classified into two distinct groups,A and B,consisting of 87 and 34 members,respectively.Besides the difference in gene number,the length of conserved DUF1618 motifs of groups A and B is also different(Figure S1).The length of DUF1618 motif in group B ranges from 134 to 199 aa,which is longer than that of group A(56–163 aa).Each group was further divided into two clades,namely A1 and A2 or B1 and B2(Figure 1).

Figure 1.Phylogenetic relationships of the identified 121 DUF1618 proteins in riceThe unrooted tree was constructed by alignments of the DUF1618 domain sequences using the software MEGA 4.0.The sequential numbers of the DUF1618 proteins in this study are shown in parentheses.The DUF1618 family in rice is classified into two groups(A and B,separated by solid line),and each group consists of two clades(A1 and A2;B1 and B2,separated by dotted lines).The DUF1618 genes with expression levels higher than 1,000 in Affymetrix GeneChip Rice Genome Array assay are marked by empty or filled stars,and six of them(indicated by filled stars)were selected for further expression analysis.

Figure 2.Chromosomal locations of the 121 rice DUF1618 genesTo simplify the figure,only the sequential numbers of the genes are shown.The different colors of the protein numbers indicate that they belong to different clades in the phylogenetic tree:clades A1(red),A2(blue),B1(black),and B2(green).Black dots indicate the centromeric positions.
Chromosomal distribution and structure of DUF1618 genes in rice
We analyzed these DUF1618 genes on their chromosomal locations and found that they are distributed unevenly on the chromosomes(Figure 2,Table S2).For example,chromosomes 1 and 3 possess 21 and 28 DUF1618 genes,respectively,but chromosomes 7 and 10 each have only one DUF1618 gene(Figure 2).Many of these genes are located as clusters on chromosomes.For instance,on chromosome 1,there are 10 DUF1618 genes located in a region of 291 kb,and on chromosome 11,10 DUF1618 genes clustered within a region of 53 kb.Two DUF1618 genes,#2 and#83,have two and three identical copies,respectively,whereas#80,#81,and#82 each possesses a total of seven copies with high nucleotide sequence identities(99–100%),and they are located as clusters.Notably,one DUF1618 protein(#64,Os05g0250101)contains two DUF1618 domains.In addition,seven DUF1618 proteins contain other conserved functional domains,such as an F-box,kinase domain,transmembrane segment,and so on(Figure 3).
DUF1618 genes in other plant species
The completion of genome sequencing in model plants and expressed sequence tag(EST)sequences of many non-model plants enables us to analyze DUF1618 gene family members in these plant species.Thirty-five plant EST databases and six plant genome sequence databases were screened(Table S1),including those of maize(Zea mays),barley(Hordeum vulgare),sorghum(Sorghum bicolor),Arabidopsis(Arabidopsis thaliana),soybean(Glycine max),and poplar(Populus×canadensis).Interestingly,the DUF1618 gene family is found only in four monocot species and not in any of the 22 analyzed dicot and other nine monocot plant species.In the genome databases of sorghum,maize,and barley,60,14,and 18 non-redundant DUF1618 family members were detected,respectively(Table 1).
We conducted phylogenetic analysis with the DUF1618 proteins,showing that all DUF1618 proteins from sorghum,maize,and barley could be integrated into the four clades of the rice phylogenetic tree(Figure 4).Most DUF1618 proteins from sorghum(46/60)and barley(11/18)were located in the A group,and all 14 DUF1618 proteins from maize were grouped into the A1 clade(Figure 4,Table 1).

Figure 3.Eight rice DUF1618 proteins contain other putative conserved domainsConserved domains are represented by colored boxes and symbols with explanation.Pkinase_tyr,protein tyrosine kinase domain;Auxin_BP,auxin-binding partner domain;Retrotrans_-gag,retrotransposon gag protein domain;BSD,BTF2-like transcription factors,synapse-associated and DOS2-like protein domain.
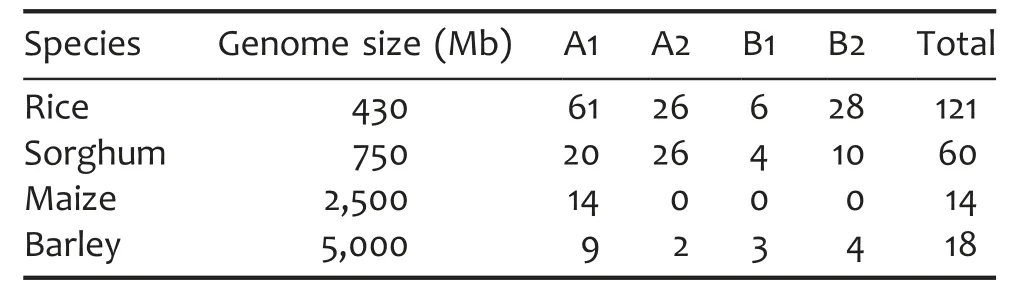
Table 1.The distribution of DUF1618 genes in the clades in four monocot species
Expression profiles of rice DUF1618 genes and the predicted protein subcellular localization
We investigated the expression profile of the 121 rice DUF1618 genes in seedling,root,leaf,stem,young panicle,and stamen,using a public expression database from the Affymetrix GeneChip Rice Genome Array(Wang et al.2010).The results showed that most of the genes were expressed at relatively low levels in the tested tissues/organs(Table S2).Only 13 DUF1618 genes(#25,#26,#35,#41,#51,#54,#59,#80,#81,#92,#112,#113,and#116)showed higher hybridization signals(>1,000)in at least one tissue/organ(Figure 1,Table S2).Among those 13 DUF1618 genes,5(#54,#59,#112,#113,and#116)belong to the A1 clade,3(#35,#41,and#51)belong to the A2 clade,and 5(#25,#26,#80,#81,and#92)belong to the B2 clade,but none is in the B1 clade.Os12g39970(#112)had the highest expression levels(6,405–14,655),similar to those of Actin 1(10,000–15,000 in different tissues).In addition,we searched these DUF1618 genes in three other microarray databases(GSE6901,GSE3053,and GSE4438).We found that the majority of the genes with relatively high expression levels are common among these databases(Figure 5A).Next,we selected six of them(#26,#35,#51,#59,#112,and#113)belonging to the A1,A2,and B2 clades(Figure 1;Table S2)to investigate their transcriptional profiles in different cultivars and tissues using quantitative reverse transcription-polymerase chain reaction(qRT-PCR).Our data showed that the expression level of these genes varied among different tissues(Figure 5B)and between indica and japonica cultivars(Figure 6).For example,#26,#35,and#113 were expressed specifically in the leaf of japonica rice Taichung 65(T65),while the#51 and#112 were expressed preferentially in the leaf of indica rice 93-11(Figure 6).In contrast,#59 and#112 had relatively high expression in the root of T65(Figure 6).
We used the Plant-PLoc server(Chou and Shen 2007)to examine the subcellular localization of the rice DUF1618 proteins,and the majority of the DUF1618 family members(81/121)were predicted to be targeted to the chloroplasts,with the others to be targeted to the cytoplasm,the mitochondria,the chloroplasts,the plasma membrane,the endoplasmic reticulum,the nucleus,the peroxisome,and the cell wall(Table S2).
Expression profiles of rice DUF1618 genes in response to abiotic stresses and salicylic acid
Based on the public microarray data,salt is the most effective signal that activates the expression of the six DUF1618 genes(#26,#35,#51,#59,#112,and#113)in an indica cultivar IR64(Figure 5C).Furthermore,we investigated the transcriptional responses of the six selected genes in seedlings of an indica line(TISL5),with different treatments.The qRT-PCR assays showed that five DUF1618 genes,except for#113,responded to cold stress;and five DUF1618 genes,except#112,responded to salt stress(Figure 7).By contrast,only one DUF1618(#112)responded to heat stress and two DUF1618s(#51 and#59)responded to drought stress(Figure 7).Salicylic acid(SA)induces the accumulation of pathogenesis-related proteins and is associated with disease resistance in plants(Koornneef and Pieterse 2008).To test whether DUF1618 family members may contribute to plant defense signaling,we treated the rice seedlings with SA and tested the expression profile of the six DUF1618 genes.The results showed that SA treatment significantly downregulated the expression of#112 and#113 DUF1618 genes,suggesting that they may be related to defense responses against pathogen invasion(Figure 7).Consistent with the public microarray data,most of the selected DUF1618 genes responded to salt stress in TISL5(Figures 5C,7).
DISCUSSION
DUF1618 is a newly identified gene family in higher plants.In this study,we found no DUF1618 genes in the databases of 22 dicot genomes or EST libraries,suggesting that the DUF1618 family originated in an ancestral monocot plant species after the divergence between monocot and dicot species.Thus,DUF1618 is a relatively young gene family.It is notable that the A1 clade is commonly shared in the four species and all the DUF1618 members in maize are grouped into this clade(Figure 4),suggesting that the A1 clade may be the primitive subfamily in these DUF1618 gene-containing species,and the DUF1618 family in maize has been evolved slowly in comparison to that in the other three species,especially in rice.

Figure 4.Phylogenetic relationship among the DUF1618 proteins from rice(Os),sorghum(Sb),maize(Zm),and barley(Hv)The unrooted tree was generated from alignments of the DUF1618 domain sequences with the software MEGA 4.0.Groups A and B are separated by a solid line and the clades are separated by dotted lines.
In the monocot grass family(Poaceae),rice has a compact genome(430 Mb),but the genome size of other monocots,such as sorghum(750 Mb),maize(2,500 Mb),and barley(5,000 Mb),have larger genomes(Arumuganathan and Earle 1991).However,the total number of non-redundant DUF1618 genes in rice,sorghum,maize,and barley is 121,60,14,and 18,respectively(Table 1).This strongly suggests that the DUF1618 gene family has expanded relatively faster in rice,not associated with the genome size variation.Many DUF1618 members with high sequence similarity are found on the chromosomes as gene clusters,suggesting that they were derived from relatively recent gene duplication events,by which the DUF1618 family in rice has become a relatively large family.This phenomenon is also observed in the NBS-LRR R protein family and F-box family and AP2/EREBP family in plants(Cannon et al.2002;Jain et al.2007;Sharoni et al.2011).
The expression analysis based on the microarray data showed that the expression levels of most DUF1618 genes(90%)are very low,suggesting that some of them may not be active genes.Of 12 high-expressing genes,most of them(10 genes)were common among these databases(Figure 5A),which supports that 6 out of the 10 common genes may cover major functional genes of the DUF1618 family.By investigation of the transcriptional levels of these six representative DUF1618 genes,we revealed the tissue specificity of DUF1618 genes in different cultivars(Figures 5B,6)and found that some DUF1618 genes are regulated by multiple stresses,especially by salinity stress(Figures 5C,7),suggesting that these genes may be important for biotic and abiotic stresses.We realized certain inconsistencies between microarray data and our qRT-PCR experimental results.These could be due to the different genotype of rice cultivars,developmental stages and/or individual treatments.Moreover,most of the DUF1618 proteins(81/121)are predicted to target to the chloroplast(Table S2),suggesting that many DUF1618 proteins may be involved in chloroplast biogenesis and/or photosynthesis pathways.The presence of a relatively large number of DUF1618 genes in the rice genome and the responses of some members to multiple stresses may indicate that the DUF1618 families important for the fitness of this species in response to stresses and in reproduction.The large number of DUF1618 family members may provide a backup strategy in case of dysfunction of an important gene,for rice reproduction and survival of multiple stresses.
Taken together,this study provides a general overview of the DUF1618 family in rice and clues for further investigation of the functions and evolutionary significance of this young gene family.
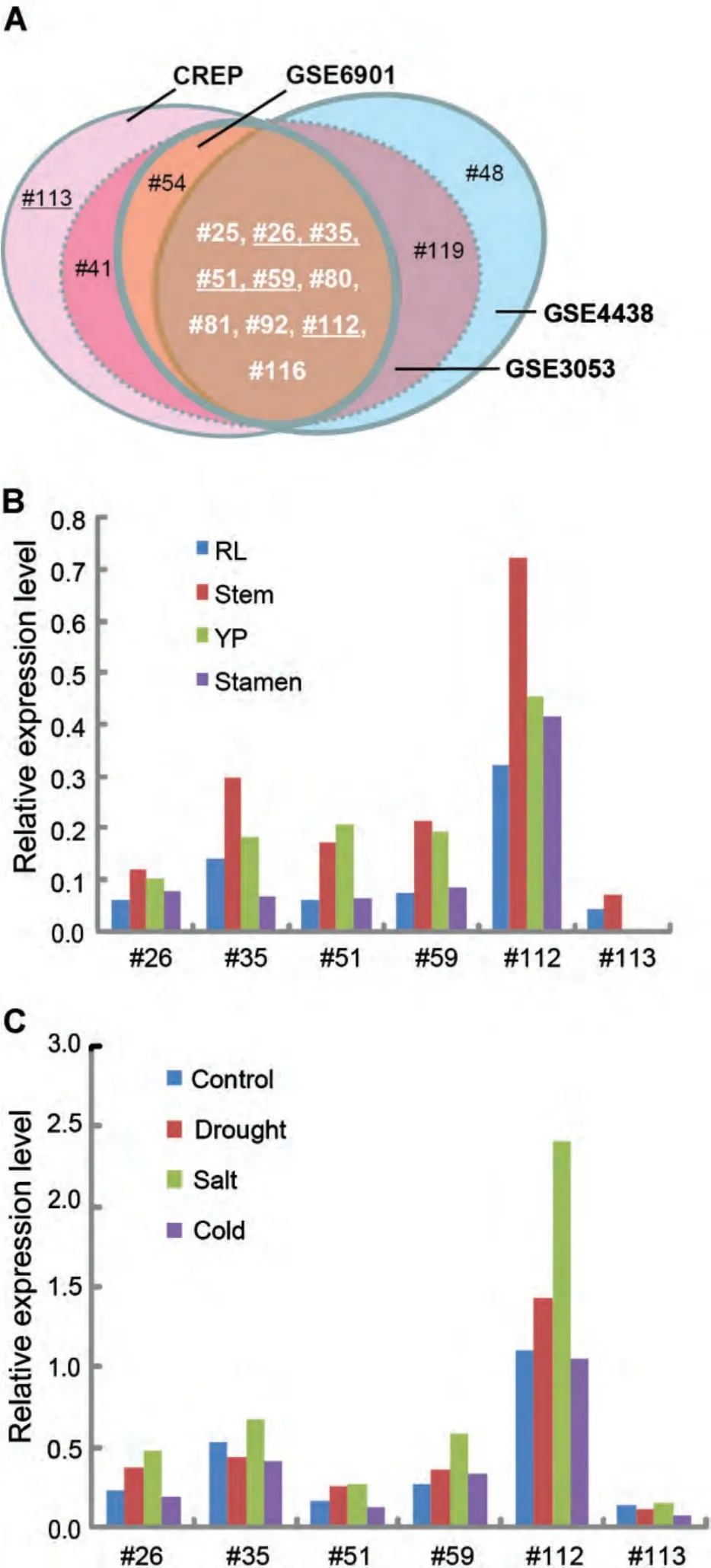
Figure 5.In-depth analysis of public expression microarray databases(A)DUF1618 genes with relative high expression were selected from four independent microarray databases.Each colored oval stands for high-expressing genes from a database.CREP and GSE3053 contain 13 high-expressing genes;GSE4438 and GSE6901 have 12 and 11 such genes,respectively.Among these four datasets,10 genes are common.(B)The expression patterns of six representative DUF1618 genes are shown based on the tissue specificity of rice of Zhenshan 97 and Minhui 63 in the CREP database.RL,root and leaf.YP,young panicles.(C)The expression profiles of six representative DUF1618 genes of IR64 in response to drought,salt,and cold stresses are presented based on the GSE6901 database.
MATERIALS AND METHODS
Database searches
The annotated DUF1618 proteins in the rice genome were searched in the NCBI protein data library.Non-redundant DUF1618 proteins with confidence by SMART/PFam(http://smart.embl-heigelberg.de/)were checked individually.The gene sequences and the DUF1618 loci were searched by NCBI BLAST(http://blast.ncbi.nlm.nih.gov/Blast.cgi).Subcellular localizations of the rice(Oryza sativa L.)DUF1618 proteins were predicted using the Plant-PLoc server(http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
The expression profiles of the 121 rice DUF1618 genes in the seedling,the root,the leaf,the stem,the young panicle,and the stamen of indica cultivars were investigated based on the public expression database CREP(Wang et al.2010;http://crep.ncpgr.cn/).In addition,three microarray databases with different varieties were used for bioinformatic analysis:GSE6901 based on indica variety IR64(Jain et al.2007;Jain and Khurana 2009;http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE6901),GSE3053 based on indica varieties IR29 and FL478(Walia et al.2005;http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3053),GSE4438 based on indica variety IR29 IR63731,and japonica cultivar M103 and Agami(Walia et al.2007;http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4438).
Phylogenetic analysis
The conserved amino acid sequence alignment of DUF1618 domains was performed by ClustalW of the software MEGA4.0.Unrooted phylogenetic trees were constructed by the neighbor-joining method with MEGA 4.0 using the DUF1618 conserved domain amino acid sequences(Tamura et al.2007).Bootstrap values were calculated from 1,000 replicates.
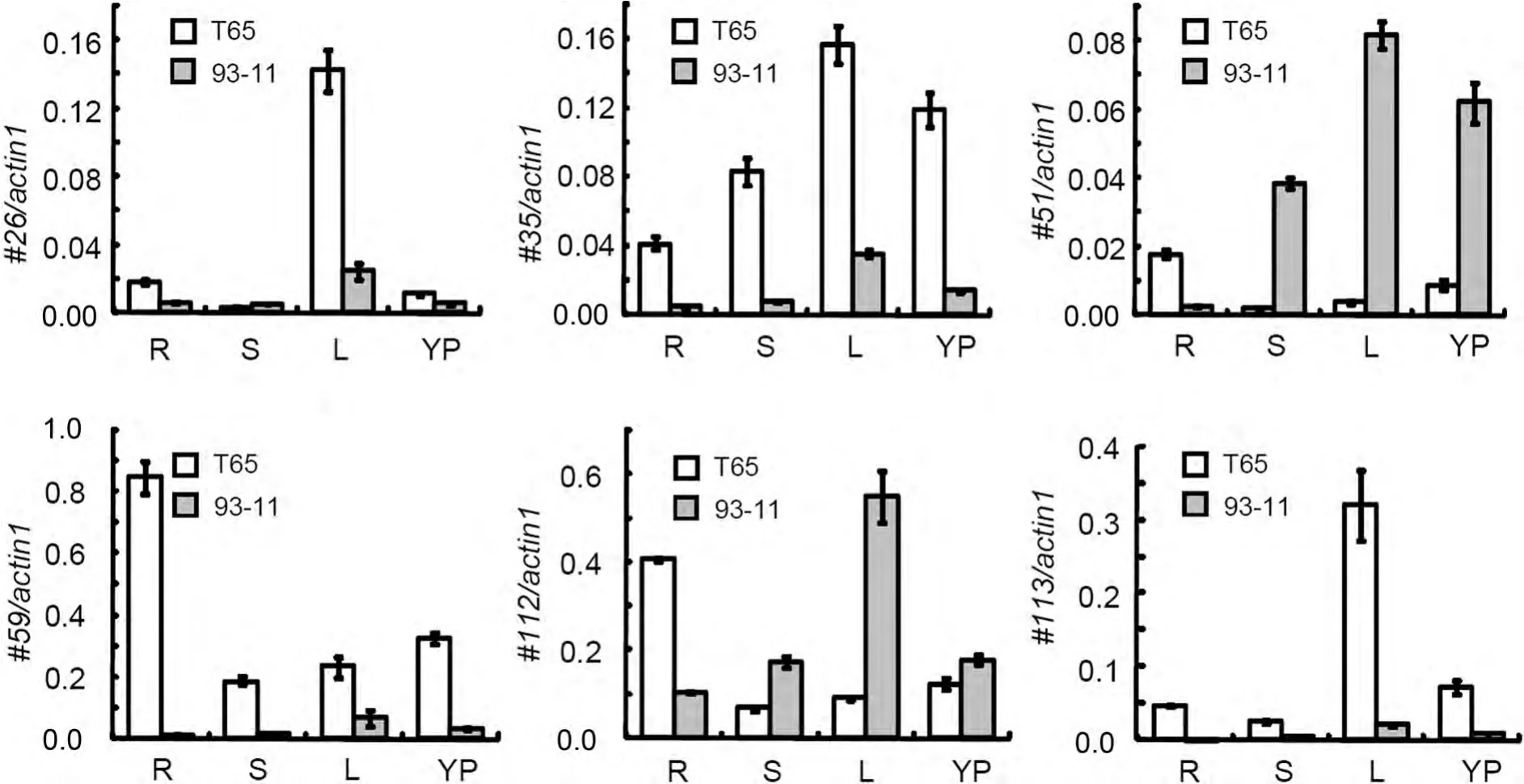
Figure 6.Validation of the expression of six DUF1618 genes by quantitative reverse transcription-polymerase chain reaction(qRT-PCR)in different tissues of riceThe expression levels were normalized by OsActin1 expression and the data shown are means±SD based on three replicates(n=3).T65 and 93-11 are japonica and indica cultivars,respectively.R,root of seedlings;L,leaf of seedlings;S,stem of seedlings;YP,young panicles.

Figure 7.Transcriptional responses of six DUF1618 genes to abiotic stresses and salicylic acid(SA)The transcriptional levels of the genes in response to abiotic stresses(cold,heat,salt,and drought)and salicylic acid(SA)were tested by quantitative reverse transcription-polymerase chain reaction(qRT-PCR)with a japonica line TISL5.The data are shown as mean ± SD(n=3).The transcriptional difference between mock and treatments was examined by Student’s t-test.Single asterisk indicates 0.01<P<0.05;double asterisks indicate P<0.01.
RNA isolation and RT-PCR analysis
The indica cultivar 93-11 and japonica cultivars Taichung 65 and TISL5(Zhang and Zhang 2001)were grown in the farm of South China Agricultural University in China.Abiotic stress treatments of rice seedlings were carried out as previously described(Li et al.2011).Total RNAs were isolated using TRIzol reagent(Invitrogen Carlsbad,CA,USA).The first strand of cDNA was synthesized from 1 μg of total RNA using the SuperScript II reverse transcription kit(Invitrogen).The qRT-PCR was conducted with gene-specific primers(Table S3).The PCRs were carried out with 30 cycles of 94 °C for 15 s,58 °C for 60 s,and 72°C for 30 s.
ACKNOWLEDGEMENTS
This research was supported by the National Natural Science Foundation of China(30800597),the Natural Science Foundation of Guangdong Province(8451064201001015),and the New Teacher Foundation of Chinese Colleges and Universities(20094404120018).We thank Dr.Zhang Q for help with stress treatment of rice samples.
Arumuganathan K,Earle E(1991)Nuclear DNA content of some important plant species.Plant Mol Biol Rep 9:208–218
Bakolitsa C,Bateman A,Jin KK,McMullan D,Krishna SS,Miller MD,Abdubek P,Acosta C,Astakhova T,Axelrod HL,Burra P,Carlton D,Chiu HJ,Clayton T,Das D,Deller MC,Duan L,Elias Y,Feuerhelm J,Grant JC,Grzechnik A,Grzechnik SK,Han GW,Jaroszewski L,Klock HE,Knuth MW,Kozbial P,Kumar A,Marciano D,Morse AT,Murphy KD,Nigoghossian E,Okach L,Oommachen S,Paulsen J,Reyes R,Rife CL,Sefcovic N,Tien H,Trame CB,Trout CV,van den Bedem H,Weekes D,White A,Xu Q,Hodgson KO,Wooley J,Elsliger MA,Deacon AM,Godzik A,Lesley S,Wilson IA(2010)The structure of Jann_2411(DUF1470)from Jannaschia sp.at 1.45 A resolution reveals a new fold(the ABATE domain)and suggests its possible role as a transcription regulator.Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1198–1204
Bateman A,Coggill P,Finn RD(2010)DUFs:Families in search of function.Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1148–1152
Cannon SB,Zhu H,Baumgarten AM,Spangler R,May G,Cook DR,Young ND(2002)Diversity,distribution,and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies.J Mol Evol 54:548–562
Chou KC,Shen HB(2007)Large-scale plant protein subcellular location prediction.J Cell Biochem 100:665–678
Dlakic M(2006)DUF283 domain of Dicer proteins has a doublestranded RNA-binding fold.Bioinformatics 22:2711–2714
Jain M,Khurana JP(2009)Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice.FEBS J 276:3148–3162
Jain M,Nijhawan A,Arora R,Agarwal P,Ray S,Sharma P,Kapoor S,Tyagi AK,Khurana JP(2007)F-box proteins in rice.Genome-wide analysis,classification,temporal and spatial gene expression during panicle and seed development,and regulation by light and abiotic stress.Plant Physiol 143:1467–1483
Karras GI,Kustatscher G,Buhecha HR,Allen MD,Pugieux C,Sait F,Bycroft M,Ladurner AG(2005)The macro domain is an ADP-ribose binding module.EMBO J 24:1911–1920
Koornneef A,Pieterse CM(2008)Cross talk in defense signaling.Plant Physiol 146:839–844
Li X,Wang C,Nie P,Lu X,Wang M,Liu W,Yao J,Liu Y,Zhang Q(2011)Characterization and expression analysis of the SNF2 family genes in response to phytohormones and abiotic stresses in rice.Biol Plantarum 55:625–633
Ohta T(2000)Evolution of gene families.Gene 259:45–52
Ohyanagi H,Tanaka T,Sakai H,Shigemoto Y,Yamaguchi K,Habara T,Fujii Y,Antonio BA,Nagamura Y,Imanishi T,Ikeo K,Itoh T,Gojobori T,Sasaki T(2006)The Rice Annotation Project Database(RAP-DB):Hub for Oryza sativa ssp.japonica genome information.Nucleic Acids Res 34:D741–D744
Qin H,Chen F,Huan X,Machida S,Song J,Yuan YA(2010)Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein–protein interaction.RNA 16:474–481
Romling U,Simm R(2009)Prevailing concepts of c-di-GMP signaling.Contrib Microbiol 16:161–181
Sharoni AM,Nuruzzaman M,Satoh K,Shimizu T,Kondoh H,Sasaya T,Choi IR,Omura T,Kikuchi S(2011)Gene structures,classification and expression models of the AP2/EREBP transcription factor family in rice.Plant Cell Physiol 52:344–360
Tamura K,Dudley J,Nei M,Kumar S(2007)MEGA4:Molecular Evolutionary Genetics Analysis(MEGA)software version 4.0.Mol Biol Evol 24:1596–1599
Tong H,Leasure CD,Hou X,Yuen G,Briggs W,He ZH(2008)Role of root UV-B sensing in Arabidopsis early seedling development.Proc Natl Acad Sci USA 105:21039–21044
Walia H,Wilson C,Condamine P,Liu X,Ismail AM,Zeng L,Wanamaker SI,Mandal J,Xu J,Cui X,Close TJ(2005)Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage.Plant Physiol 139:822–835
Walia H,Wilson C,Zeng L,Ismail AM,Condamine P,Close TJ(2007)Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage.Plant Mol Biol 63:609–623
Wang L,Xie W,Chen Y,Tang W,Yang J,Ye R,Liu L,Lin Y,Xu C,Xiao J,Zhang Q(2010)A dynamic gene expression atlas covering the entire life cycle of rice.Plant J 61:752–766
Zhang Z,Zhang G(2001)Fine mapping of the S-c locus and markerassisted selection using PCR markers in rice.Acta Agron Sin 27:704–709
SUPPORTING INFORMATION
Additional supporting information can be found in the online version of this article:
Figure S1.Amino acid sequence alignment of some selected DUF1618 domains
(A)Proteins belong to group A,which share average 39.2%similarity in the domain sequences.(B)Proteins belong to group B,which share average 40.5%similarity in the domain sequences
Table S1.The EST databases for 35 species screened in this study
Table S2.Expression patterns and predicted localization of DUF1618 genes in rice
Table S3.Primers for qRT-PCR in this study
 Journal of Integrative Plant Biology2014年2期
Journal of Integrative Plant Biology2014年2期
- Journal of Integrative Plant Biology的其它文章
- Diversity characterization and association analysis of agronomic traits in a Chinese peanut(Arachis hypogaea L.)mini-core collection
- Why mosaic?Gene expression profiling of African cassava mosaic virus-infected cassava reveals the effect of chlorophyll degradation on symptom development
- Two hydroxypyruvate reductases encoded by OsHPR1 and OsHPR2 are involved in photorespiratory metabolism in rice
