由4-甲基-1,2,3-噻二唑-5-甲酸构筑的铜配合物的合成、晶体结构及与DNA作用
胡未极 武大令 沈金杯赵国良*,,(浙江师范大学化学与生命科学学院,金华3004)(浙江师范大学行知学院,金华3004)
由4-甲基-1,2,3-噻二唑-5-甲酸构筑的铜配合物的合成、晶体结构及与DNA作用
胡未极1武大令1沈金杯1赵国良*,1,2
(1浙江师范大学化学与生命科学学院,金华321004)
(2浙江师范大学行知学院,金华321004)
由4-甲基-1,2,3-噻二唑-5-甲酸(HMTC,C4H4N2O2S)和菲咯啉(Phen)合成了2个铜配合物[Cu(MTC)2(H2O)2]n(1),[Cu2(MTC)2(Phen)2(H2O)4](MTC)2(2)。用元素分析、红外光谱、热重分析、粉末X射线衍射进行表征,用单晶X射线衍射方法测定了配合物的晶体结构。结果表明,配合物1是一维链状结构,属于单斜晶系,P21/c空间群,中心金属铜(Ⅱ)离子的配位构型是一个畸变的四方锥结构。配合物2属于三斜晶系,P1空间群,是一个双核结构,由2个配位水分子上的氧桥连2个铜(Ⅱ)离子形成六配位的扭曲八面体结构。用溴化乙锭荧光探针测定了配体和配合物与DNA的相互作用,结果显示无论是配体还是配合物均能使EB-DNA复合体系的荧光发生不同程度的猝灭,且配合物的作用强度大于配体,具有刚性平面辅助配体的配合物2的作用强度又大于不加辅助配体的配合物1。
铜配合物;4-甲基-1,2,3-噻二唑-5-甲酸;晶体结构;DNA作用
In recent years,the rational design and synthesis of functional complexes construction based on organic ligands have attracted considerable attention because of the novel topological structures[1]and potential application in gas adsorption[2],separation[3],catalysis[4], magnetism[5],luminescence[6]and bioactivity[7].
Copper is an indispensable micronutrients to human,which is closely related to human health and plays an important role in metabolism,synthesizing DNAandsoon[8-9].Inthe previous reports,the interaction between polypyridine transition metal complexes and DNA has drawn extensive attention[10-11].Numerous studies concluded that polypyridine copper complexes possessed plentiful potential bioactivity due to the intercalation of the complexes into DNA,such as the work done by Sigman[12],Thomas[13]and Reddy[14].
Thiadiazol derivatives are widely used in the area ofpesticidesandpharmaceuticsowingtotheir extensivebiologicalactivityincludingantifungal, antibacterial,antitubercular,antimycobacterial,anticancer,diuretic,and hypoglycemic properties[15-17].So it is significant to carry out the research concerning mechanisms and bonding abilities between copper thiadiazol carboxylate polypyridine complexes and DNA,which can help us to design and synthesize DNA secondary structure probes,nucleic acid location reagents and anti-cancer drugs.
Amongthiadiazolcompounds,biological activities of 1,3,4-thiadiazol compounds and 1,2,3-thiadiazol compounds have been reported for many times,while reports about boiogical activity based on 1,2,3-hiadiazol derivatives haven′t been seen before. Inthispaper,wereportthesynthesisand characterizations of two new copper(Ⅱ)complexes constructed by 4-methyl-1,2,3-thiadiazol-5-carboxylic acidandphenanthroline.Theinteractionsof complexes and ligand with DNA were also studied by EtBr fluorescence probe.
1 Experimental
1.1Materials and measurements
Allreagentsandsolventsusedwereof commercially available quality and without purified before using.FTIR spectra were recorded on a Nicolet NEXUS 670 FTIR spectrophotometer using KBr discs in the range of 4 000~400 cm-1.Elemental analyses of C,H,N were performed on elemental analyzer,Elementar Vario ELⅢ.Crystallographic data were collected on a Bruker Smart ApexⅡCCD diffracto-meter.A Mettler Toledo thermal analyzer TGA/SDTA 851ewas used to carry out the thermoanalytical analysis with a heating rate of 10℃·min-1from 30 to 600℃in air atmosphere.Powder X-ray diffraction(PXRD)data were collected on a PW 3040/60 Focus X-Ray diffractometer using Cu Kα radiation(λ=0.154 06 nm, 10°·min-1,1°~30°).Fluorescence spectra were measured at room temperature with an Edinburgh FL-FS 920 TCSPC system.
1.2Synthesis of[Cu(MTC)2(H2O)2]n(1)
A mixture of HMTC(0.288 g,2.0 mmol),CuCl2·2H2O(0.170 g,1.0 mmol),NaOH(0.040 g,1.0 mmol) and H2O(15 mL)was put into a 50 mL vessel and stirred for 8h at room temperature.The resulted solution was allowed to evaporation at room temperature for two weeks.Green crystals suitable for singlecrystal analysisandphysicalmeasurementswere obtained,washed with distilled water,and dried in air.Yield:58%(based on HMTC).Anal.Calcd.for C8H10N4O6S2Cu(%):C,24.88;H,2.59;N,14.51.Found (%):C,24.79;H,2.42;N,14.42.IR(KBr,cm-1):3 300, 1 631,1 500,1 448,1 390,1 370,1 284,1 218,1 031, 784,733,597,534.
1.3Synthesisof[Cu2(MTC)2(Phen)2(H2O)4](MTC)2(2)
A mixture of HMTC(0.144 g,1 mmol),CuCl2·2H2O(0.085 g,0.5 mmol),NaOH(0.040 g,1.0 mmol), Phen(0.100 g,0.5 mmol)and EtOH(5 mL)/H2O(5 mL)was put into a 50 mL vessel and stirred for 8 h at room temperature.The resulted solution was allowed to evaporation at room temperature for two weeks. Green crystals suitable for single-crystal analysis and physical measurements were obtained,washed with distilled water,and dried in air.Yield:50%(based on HMTC).Anal.Calcd.for C40H36N12O12S4Cu2(%):C,42.40; H,3.18;N,14.84.Found(%):C,42.21;H,3.07;N, 14.72.IR(KBr,cm-1):3 521,3 061,1 589,1 512,1 440,1 212,1 149,1 110,1 024,874,785,738,590.
1.4X-ray diffraction analysis
The single crystals of the two complexes with approximate dimensions(0.360 mm×0.150 mm×0.100 mm,1;0.266 mm×0.238 mm×0.069 mm,2)were selected and mounted on a Bruker Smart ApexⅡCCD diffractometer.The diffraction data were collected using a graphite monochromated Mo Kα radiation(λ= 0.071 073 nm)at 296(2)K.The employed single crystals exhibit no detectable decay during the data collection.Absorption corrections were applied using SADABS[18].The structure was solved by direct methods with the SHELXS-97[19]program package and refined with the full-matrix least-squares technique based on F2using the SHELXTL-97[20]program package. Hydrogen atoms on water molecules were located in a difference Fourier map and included in the subsequent refinement using restrains(OH)0.085 nm,HH 0.13 nm)with Uiso(H)=1.5 Ueq(O).Other hydrogen atoms were added theoretically.Detail information about the crystal data is summarized in Table 1.Selected interatomic distances and bond angles are given in Table 2~3.All hydrogen bonds are given in Table 4.
CCDC:825716,1;1435439,2.

Table 1Crystallographic data for the complexes 1 and 2

Table 2Selected bond distances(nm)and angles(°)of the complex 1

Continued Table 1

Table 3Selected bond distances(nm)and angles(°)of the complex 2

Table 4Hydrogen bond distances(nm)and bond angles(°)of the complexes 1 and 2
1.5EB-DNA binding study by fluorescence spectrum
TheinteractionsoftheHMTCligandand complexes with calf thymus DNA(CT-DNA)were studied by ethidium bromide(EB)fluorescent probe. The experiment was carried out by adding different volumes of compound solution(10-4mol·L-1for HMTC ligand or complexes)to a 10 mL colorimetric cylinder prepared 2 h in advance,which contained 1.0 mL 200 μg·mL-1EB,1.0 mL 200 μg·mL-1CT-DNA,and 2.0 mL tris-HCl buffer solution(pH=7.4),then the mixed solution were diluted with double-distilled water.The final solutions were incubated for 12 h at 4℃.The fluorescence was recorded at an excitation wavelength of 251 nm and emission wavelength between 520 nm and 700 nm.
2 Results and discussion
2.1Crystal structure analysis of the complexes
2.1.1Structure analysis of complex 1
Single-crystalX-rayanalysisshowsthatthe complex 1 crystallizes in the monoclinic system with space group P21/c and Z=4.Each asymmetric unit consists of one Cu2+,two MTC ligands,two coordinated water.The Cu(Ⅱ)ion is five-coordinated with four O atoms from two coordinated water(O1W,O2W)and two ligands(O1,O3i)and one N atom from the ligand (N4)(Fig.1).The O2W,O1,O3iand N4 are locted in thebasalplane,whereastheO1Wfromthe coordinated water occupied the axial position,forming a distorted pentahedron coordinated geometry.The distance of Cu-O is 0.194 6 nm to 0.223 7 nm,andCu-N is 0.204 3 nm,according with the distance of Cu-O and Cu-N in the literature[21-22].

Fig.1(a)Ellipsoidal structural view of 1 with probability level of 30%;(b)View of the coordination environment of Cu(Ⅱ)ion for 1
This dual-nuclei is bridged by one ligand forming a 1D chain along the a axis.There are obviously face to face π-π stacking interactions in 1(Fig.2),with the center to center distance of 0.378 3 nm and 0.351 0 nm between the aromatic rings and the adjacent unit.

Fig.2π-π packing interactions in complex 1
Intermolecular hydrogen bonding interactions are observed in 1(Fig.3).Both of hydrogen bonding interactions and the π-π stacking interactions made the molecules further connect into a 3D supramolecular network(Fig.4).
2.1.2Structure analysis of complex 2

Fig.3Hydrogen bonding interactions of complex 1

Fig.4Three-dimensional supramolecular network of complex 1

Fig.5(a)Ellipsoidal structural view of 2 with probability level of 30%;(b)View of the environment of Cu(Ⅱ)ion for 2
Single-crystalX-rayanalysisshowsthatthe complex 2 crystallizes in the triclinic system with space group P1 and Z=1.Complex 2 displays a binuclear structure(Fig.5),which is bridged by two oxygen atoms from two coordinated water.Each asymmetric unit consists of two Cu2+ions,two MTC ligands,two Phen ligands,four coordinated water and two free MTC anions.Each Cu2+ion adopts six-coordinated mode with a distorted octahedral geometry,by two N atoms(N5,N6)from one Phen and four oxygen atoms (one from the MTC ligand O1,the another three from three coordinated water O1W,O2W,O1Wi).The O1, O1Wi,O2W,N5 are locted in the equatorial plane, and the O1W and N6 occupy the axial positions.The distance of Cu-O is 0.196 9 nm to 0.277 5 nm and Cu-N is 0.200 7 nm to 0.202 1 nm,according with the distance of Cu-O and Cu-N in the literature[23-24].
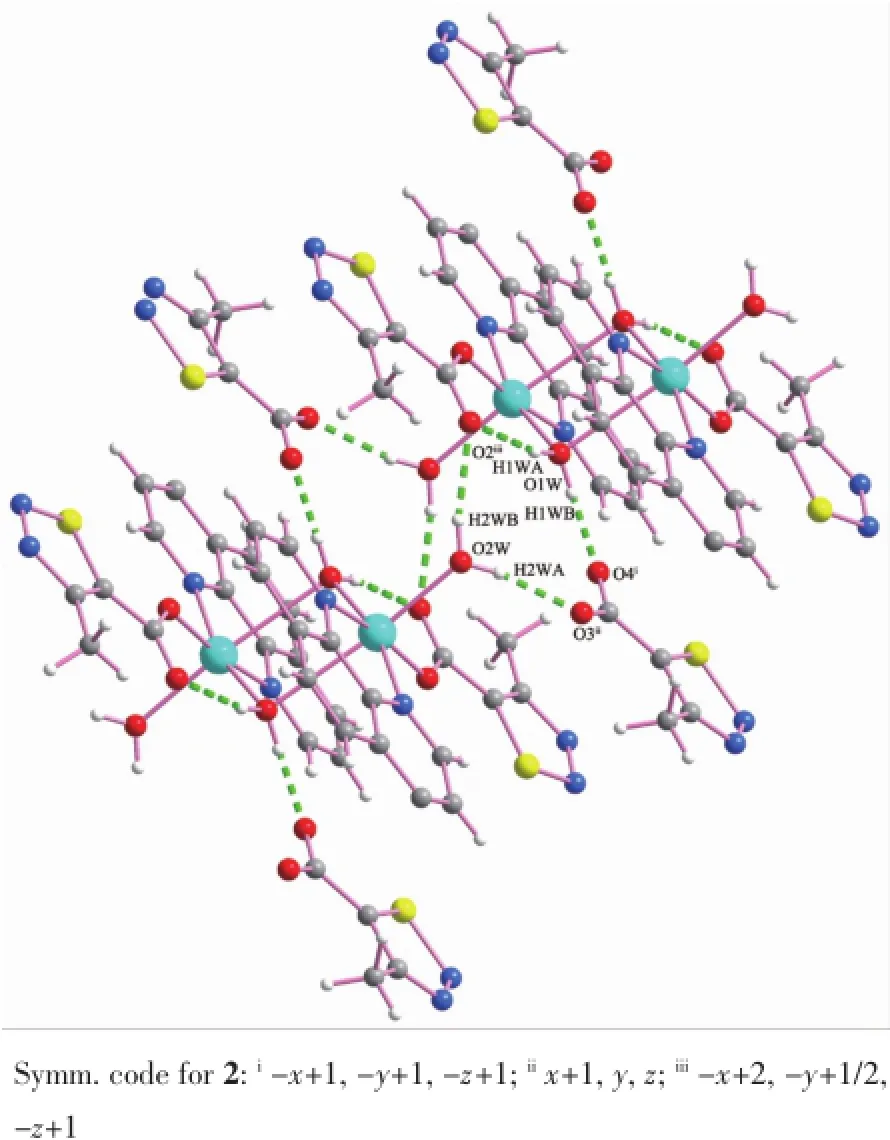
Fig.6Hydrogen bonding interactions of complex 2
As shown in Fig.6,in the complex2,the coordinated water molecules(O1W and O2W)act as hydrogen donors,contributing hydrogen atoms(H1WA, H1WB and H2WA and H2WB)to O4i,O3ii,O2iiiand O2 forming the hydrogen bonds.The structure is extended into a 2D network through the intermolecular hydrogen bonding interactions(H1WA…O2 0.184 nm, H1WB…O4i0.170 nm,H2WA…O3ii0.187 nm, H2WB…O2iii0.202 nm).
We have analyzed the PXRD pattern and the simulated powder patterns form the single crystal data of 1 and 2 to examine the phase purity of the complexes.As shown in Fig.7,it is indicated that the complexes 1 and 2 have highly crystalline purity.
2.3IR spectrum
In the IR spectrum of complex 1,there are two strong peaks near 3 300 cm-1(ν(O-H))and 597 cm-1(δ(O-H)),which corresponds to presence of coordinated water molecules.The peak at 1 500 cm-1may be assigned to the asymmetric COO-stretching vibration, the peak at about 1 390 cm-1are associated with the symmetric COO-stretching vibrations,in stead of the features at 1 733 cm-1associated with the COO-stretching vibrations of the ligand.It is indicated that the coordination of the carboxylate oxygen atoms with the Cu2+ions[25].For complex 2,the peaks at 3 521cm-1and 570 cm-1show the presence of coordinated water molecules,two absorption peaks at 1 512 cm-1(ν(OCO)asym)and 1 440 cm-1(ν(OCO)sym)indicate the coordination of the carboxylate oxygen atoms with the Cu2+ions.Two bands occurring at 1 592 cm-1(ν(C= N)),861 cm-1(δ(C-H))show the coordination of nitrogen atoms from Phen[26].These results are consistent with the X-ray diffraction structural analysis.
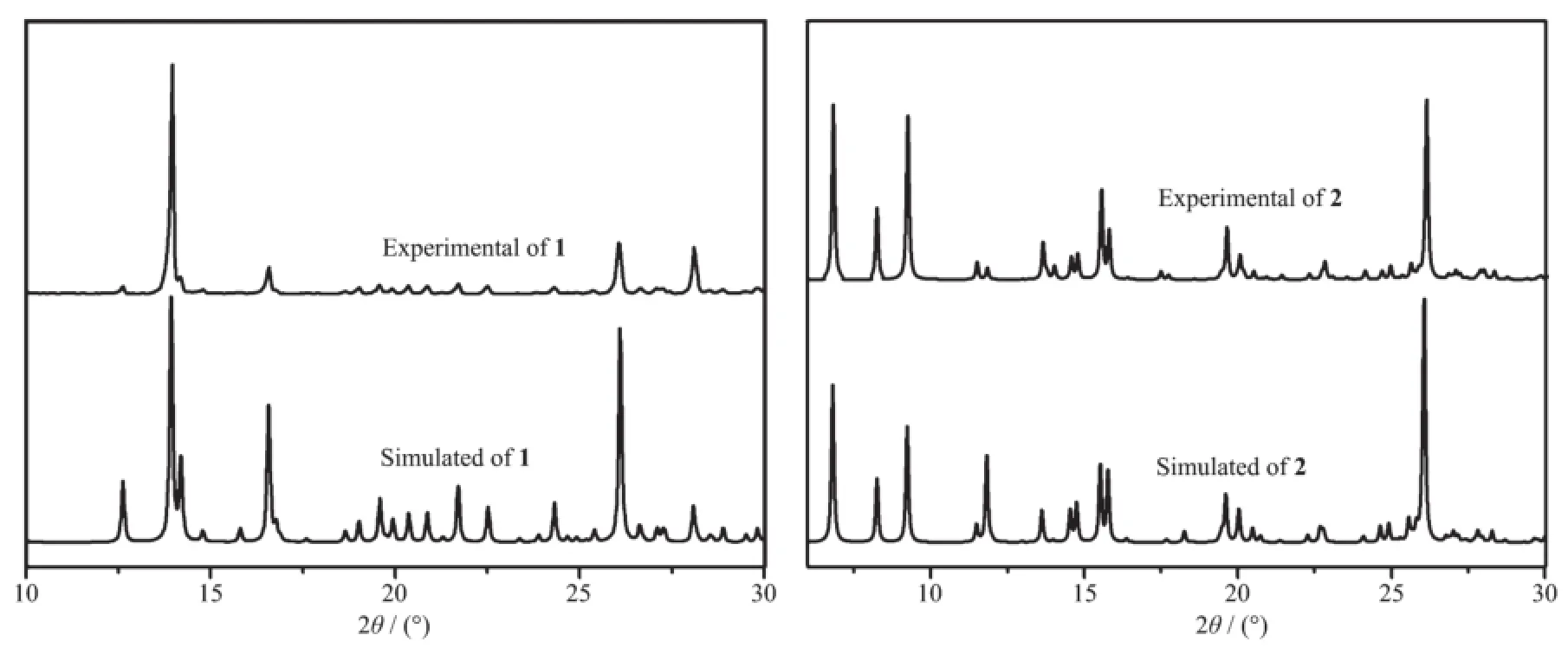
Fig.7 Experimental and simulated PXRD patterns for complexes 1 and 2
2.4Thermo-gravimetric analysis
As depicted in Fig.8,for 1,the first weight loss of 10.02%up to 194℃can be assigned to the loss of the two water molecules(Calcd.9.33%).The second weight loss of 74.46%up to 550℃can be assigned to the loss of the two ligands(Calcd.74.22%),then, the residue weight of 15.52%corresponds to CuO (Calcd.16.45%).For 2,the first weight loss of 6.56% up to 103℃can be assigned to the loss of the four water molecules(Calcd.6.36%).The second weight loss of 82.38%up to 450℃can be assigned to the loss of the two Phen molecules and the four ligands (Calcd.82.66%),then,the residue weight of 11.26% corresponds to CuO(Calcd.10.98%).
2.5EB-DNA binding study
1.当前在小学生英语的教学领域,有一大部分教师沿用传统英语教学方法,不仅在方法上被时代所淘汰,在教学理念上也非常陈旧和落后,很多农村的教师在新式的教学方法方面几乎没有涉及过信息化教学模式,在他们的教学过程中根本无法有效地在课堂上吸引学生的注意力。在这种情况下,学生学习积极性很难被充分调动。
Fig.9 shows the emission spectra of EB bounded to DNA with HMTC ligand,complexes.As concentrations of the compounds increasing,the emission intensity at 592 nm of EB-DNA system decreased in different degrees.According to the classical Stern-Volmer equation[27]:I0/I=1+Ksqr,where I0and I represented the fluorescence intensities in the absence or presence of the compounds,respectively.r is the concentration ratio of the compounds to DNA.Ksqis a linear Stern-Volmer quenching constant.The Ksqvalue is obtained as the slope of I0/I versus r linear plot.

Fig.8TG curves of complexes 1 and 2

Fig.9Emission spectra of EB-DNA system in the absence and presence of HMTC ligand,1 and 2
From the inset in Fig.9,the Ksqvalue are 0.240, 0.525 and 1.855 for the HMTC ligand and complexes 1 and 2.It is indicated that the interaction of the ligand itself and complexes with DNA could release some free EB from EB-DNA,because of the present of thiadiazol rings.For complex 2,it has the strongest interaction with the DNA(related to HMTC ligand and complex 1)because of releasing more free EB form EB-DNA by the big Phen planar molecules.
3 Conclusions
Two copper(Ⅱ)complexes[Cu(MTC)2(H2O)2]n(1) and[Cu2(MTC)2(Phen)2(H2O)4](MTC)2(2)based on the thiadiazol derivative HMTC have been successfully synthesized.The complex 1 exhibits extended 3D frameworkbyintermolecularhydrogenbonding interactions and π-π packing interactions,and the complex 2 displays 2D frameworks by intermolecular hydrogen bonding interactions.The complex 2 has stronger interaction with the DNA than complex 1, might be resulted from releasing more free EB form EB-DNA by the big Phen planar molecules.
References:
[1]Yaghi O M,O′Keeffe M,Ockwig N W,et al.Nature,2003, 423(6941):705-714
[2]Ocking N W,Friedrichs D,O′Keeffe M,et al.J.Am.Chem. Soc.,2007,129(7):1858-1859
[3]Gurdal Y,Keskin S.Ind.Eng.Chem.Res.,2012,51(21): 7373-7382
[4]Shultz A M,Farha O K,Hupp J T,et al.J.Am.Chem.Soc., 2009,131(12):4204-4205
[5]Guo Y N,Xu G F,Gamez P,et al.J.Am.Chem.Soc.,2010, 132(25):8538-8539
[6]Binnemans K.Chem.Rev.,2009,109(9):4283-4374
[7]DanielKG,GuptaP,HarbachRH,etal.Biochem.Pharmacol., 2004,67(6):1139-1151
[8]Zhou H,Zheng C,Zou G J,et al.Int.J.Biochem.Cell Biol., 2002,34:678-684
[9]Cai X,Pan N,Zou G.Biometals,2007,20(1):1-11
[10]Peng B,Chao H,Sun B,et al.J.Inorg.Biochem.,2007,101 (3):404-411
[11]Jiang Q,Xiao N,Shi P F,et al.Coord.Chem.Rev.,2007, 251(15/16):1951-1972
[12]Sigman D S.Acc.Chem.Res.,1986,19(6):180-186
[13]Thomas A M,Nethaji M,Chakravarty A R.J.Inorg.Biochem., 2004,98(6):1087-1094
[14]Reddy P A N,Santra B K,Nethaji M,et al.J.Inorg.Biochem., 2004,98(2):377-386
[15]Mathew,Keshavayya J,Vaidya V P,et al.Eur.J.Med. Chem.,2007,42(6):823-840
[16]Amir M,Kumar H,Javed S A.Bioorg.Med.Chem.Lett., 2007,17(16):4504-4508
[17]Karegoudar P,Prasad D J,Ashok M,et al.Eur.J.Med. Chem.,2008,43(4):808-815
[18]Sheldrick G M.SADABS,Program for Empirical Absorption Correction of Area Detector Data,University of Göttingen, Germany,1997.
[19]Sheldrick G M.SHELXS 97,Program for Crystal Structure Solution,University of Göttingen,Germany,1997.
[20]Sheldrick G M.SHELXL 97,Program for Crystal Structure Refinement,University of Göttingen,Germany,1997.
[21]WANG Ji-Xiao(王继虓),WANG Che(王澈),GAO Xue(高雪),et al.Chinese J.Inorg.Chem.(无机化学学报),2015,31 (5):923-929
[22]HU Chun-Yan(胡春燕),XIAO Wei(肖伟),TAO Bai-Long(陶白龙),et al.Chinese J.Inorg.Chem.(无机化学学报),2014, 30(2):257-263
[23]Xu T G,Xu D J.J.Coord.Chem.,2005,58(5):437-442
[24]JIN Chao(金超),WANG Yin-Ge(王银歌),QIAN Hui-Fen(钱惠芬),et al.Chinese J.Inorg.Chem.(无机化学学报),2015, 31(2):413-419
[25]Gao H L,Yi L,Zhao B,et al.Inorg.Chem.,2006,45(15): 5980-5988
[26]Singh U P,Tyagi S,Sharma C L,et al.Dalton Trans.,2002 (23):4464-4470
[27]LakowiczJR.WeberG.Biochemisty,1973,12(21):4161-4170
Syntheses,Crystal Structures and DNA-Binding of Two Copper(Ⅱ)Complexes Constructed by 4-Methyl-1,2,3-thiadiazol-5-carboxylic Acid
HU Wei-Ji1WU Da-Ling1SHEN Jin-Bei1ZHAO Guo-Liang*,1,2
(1College of Chemistry and Life Science,Zhejiang Normal University,Jinhua,Zhejiang 321004,China)
(2Xingzhi College,Zhejiang Normal University,Jinhua,Zhejiang 321004,China)
Two copper complexes[Cu(MTC)2(H2O)2]n(1)and[Cu2(MTC)2(Phen)2(H2O)4](MTC)2(2)were synthesized by 4-methyl-1,2,3-thiadiazol-5-carboxylic acid(HMTC,C4H4N2O2S)and 1,10-phenanthroline(Phen),and structurally characterized by elemental analysis,IR spectra,TG,PXRD and single crystal X-ray diffraction method.The single crystal X-ray diffraction reveals that the complex 1 formed a 1D chain coordination ploymer and crystallized in monoclinic with space group P21/c.Central Cu2+was five-coordinated forming a distorted pentahedron coordination geometry.The complex 2 was a binuclear structure,which was bridged by two oxygen atoms from two coordinated water,and crystallized in triclinic with space group P1.Central Cu2+was six-coordinated forming a distorted octahedron coordination geometry.In addition,the interaction of complexes and ligand with DNA were also studied by ethidium bromide fluorescence probe.The interaction of complex 2 with DNA was stronger than complex 1.CCDC:825716,1;1435439,2.
copper complex;4-methyl-1,2,3-thiadiazol-5-carboxylic acid;crystal structure;DNA-binding
O614.121
A
1001-4861(2016)08-1467-09
10.11862/CJIC.2016.183
2016-04-18。收修改稿日期:2016-06-29。
浙江省公益性技术应用研究计划项目(No.2014C32014)。
*通信联系人。E-mail:sky53@zjnu.cn

