Expression Characteristics and Putative Functions of KIF3A/KIF3B During Spermiogenesis of Phascolosoma esculenta
DU Chen, MU Danli , GAO Xinming, LUO Shengyu, WANG Jianping,JIN Shan, and ZHU Junquan, *
Expression Characteristics and Putative Functions of KIF3A/KIF3B During Spermiogenesis of
DU Chen1), MU Danli1), GAO Xinming1), LUO Shengyu1), WANG Jianping2),JIN Shan1), and ZHU Junquan1), *
1),,315211,2),315012,
The microtubule (MT)-associated proteins KIF3A and KIF3B are ubiquitously expressed in a wide range of taxa. This study investigated the functions of these proteins in spermiogenesis, which involves various MT-dependent processes, in. We cloned the complete cDNA of-KIF3A/3B. Structural predictions showed that-KIF3A/3B are composed of a highly conserved motor domain, a coiled-coil domain, and a tail domain. Real-time quantitative PCR showed thatare expressed in all tissues evaluated, with the highest levels in sperm masses. Fluorescencehybridization and immunofluo- rescence were employed to analyze the dynamic expression patterns of KIF3A/3B during spermiogenesis.-KIF3A/3B consistently co-localized with MTs at all stages of spermiogenesis, indicating their potential functions in cargo trafficking.-KIF3A/3B co- localized with mitochondria, suggesting that they may mediate mitochondrial movement. In late-stage spermatids and mature sperm, co-localization was detected in the midpiece, suggesting that-KIF3A/3B could facilitate midpiece formation. The co-localization of-KIF3A and-KIF3B at all stages of spermiogenesis suggests their function as the heterodimeric KIF3AB. Basing on the ob- served temporal and spatial expression patterns of-KIF3A/3B, MTs, and mitochondria in our study, we suggest that heterodimer KIF3AB has a potential role in acrosome biogenesis, sperm head remodeling, enflagellation, and mitochondrial migration during spermiogenesis inIn addition, our study on the morphological characteristics of spermatogenic cells provided fundamental data on the reproductive biology of
; spermiogenesis; kinesin 3A/3B; heterodimerization
1 Introduction
In general, spermatogenesis can be divided into three phases: a mitotic phase in which spermatogonia produce sper-matocytes, a meiotic phase in which spermatocytes produce haploid round spermatids, and a haploid phase (Rupik.,2011; O’Donnell and O’Bryan, 2014). During these phases,biochemical and morphological changes in germ cells varyconsiderably among species, especially among invertebrates. Invertebrate sperms can be divided into several groups ac- cording to their differences in the head, flagellum, and acro- some. For example, most invertebrate sperms, such as the sperms of mollusks(Dang., 2012) and(Nuurai., 2016), contain an acrosome and a long flagellum. However, the amoeboid- shaped sperms of the nematodelack the acrosome and flagellum (Singson 2001). The none-flage- llate sperms of the crustaceansandhave spiked heads (Lu., 2014; Zhao., 2017). For flagellate sperms, the integrity of the sperm tail is essential for sperm motility and fertilization (Lehti and Sironen, 2016). The midpiece is a conserved structure of sperm tails.It consists of the intermediate axoneme and peripheral mitochondria, which provide energy for sperm motility (Ramalhosantos., 2009; Amaral., 2013). The acrosome is a landmark for spermatid polarity,and it is formed by secretory vesicles from the Golgi apparatus. The acrosome has important functions in fertilization (Yang and Sperry, 2003). The acrosome biogenesis, head deformation, and tail formation of sperm are clearly microtubule (MT)- dependent processes (Moreno., 2006; O’Donnell and O’Bryan, 2014).
MTs serve as the rails for intracellular transport, ena- bling motor proteins to ‘walk along’. Enflagellation and spermatid nuclear reshaping rely on MTs and MT-asso- ciated motor proteins (Fawcett., 1971; O’Donnell and O’Bryan, 2014; Zhao., 2017). The Kinesin superfa- mily is a group of MT-dependent proteins involved in in- tracellular transport (Hirokawa and Noda, 2008). Among KIFs, Kinesin II controls anterograde trafficking (Funa-bashi., 2018).In mammals, Kinesin II genes include,,, and(Gilbert., 2018). This subfamily is ubiquitously expressed at high levels in organisms (Henson., 1997; Marszalek and Goldstein, 2000). As members of Kinesin II, KIF3A and KIF3B are expressed in various tissues and are highly expressed in the testis and nervous system (Hall., 1992; Henson., 1997; Dang., 2012; Lehti., 2013). KIF3C and KIF17 are only highly expressed in the nervous sys- tem (Yang., 2001). KIF3A/3B serve important func- tions in the anterograde transport of membrane-bound or- ganelles (Marszalek and Goldstein, 2000; Zhao., 2017) and newly synthesized proteins from the endoplasmic re- ticulum (Stauber., 2006). KIF3A/3B are also involved in intraflagellar transport (IFT).They can transport largeprotein complexes from the base to the tip of the cilia and flagella (Pan., 2006; Scholey, 2012). They also par- ticipate in cell division (Fan and Beck, 2004; Haraguchi., 2006; Kodani., 2013; Shen., 2017). Thefunctions of KIF3A and KIF3B in spermiogenesis have be-come a major focus of research. In the reptile,is highly expressed in the testis and sperma- tids (Hu., 2012). In the cephalopod, KIF3A/3B play potential roles in nuclear deformation and enflagellation (Wang., 2010; Dang., 2012). In the crustaceansand, KIF3A/3B are involved in acrosome formation and nuclear reshaping (Lu., 2014; Zhao., 2017). In sand dol- lar sperm, KIF3A/3B are located in the midpiece and fla- gellum (Henson., 1997). In,KIF3A/3B may transport mitochondria during mid- piece formation (Zhao., 2018). In murine testes, KIF3A/ 3B are involved in sperm tail formation and head shaping (Miller., 1999b; Lehti., 2013). In brief, KIF3A/ 3B are essential for spermiogenesis in various animals,especially in the sperm head deformation and tail forma- tion. However, the functions of KIF3A/3B in Sipuncula havenot been determined. Moreover, species belonging to Si-puncula developed specialized reproductive strategies. Du- ring spermatogenesis, the ribbonlike testis releases sper- matocytes into the coelomic fluid, wheregerm cells gather into cell masses and grow into mature spermatozoa. Af- ter storage in the nephridia for a brief period, fertilization in sipunculids is completed(Staton, 1994). The unique aspects of this process emphasize the importance of Sipuncula research, particularly the studies of spermio- genesis.is an economically valu- able endemic Chinese species belonging to Sipuncula (Li, 1992). Aspects of its reproductive and developmental bio- logy have been described (Zhu., 2007; Long.,2015).has a typical flagellate sperm. Tail formation and head remodeling can be observed during the spermiogenesis of. Thus, we selected this spe-ciesas a model toexplore the functions of KIF3A/3B in spermatogenesis.
In this study, 1) full-lenth KIF3A and KIF3B cDNA were cloned, 2) the expression patterns ofmRNA in various tissue types were evaluated, and 3) FISH and immunofluorescence were performed to examine the functions of-KIF3A/3Bin sperm head remodeling and tail formation. In addition, detailed descriptions of spermio- genesis inwere obtained. Our results provide fundamental data for studying the reproductive biology of.
2 Materials and Methods
2.1 Preparation of Animals and Sampling
Adultmaleused in this expe- riment was obtained from Xiangshan county of Ningbo (Zhejiang, China). Experimental animals were selected by observing the germ cells in coelomic fluid. The coelomic fluid was stratified with a centrifuge (3000×, 3min) to pu- rify the spermatid masses. Then, the sperm masses were collected from the top of the liquid. The living individuals were dissected to collect coelomic fluid, the purified sper- matid masses, intestine, nephridium, retractor muscle, and body wall. Tissues were stored at −80℃ for RNA extrac- tion. For observation of HE staining, transmission electronmicroscopy, FISH, and IF, spermatid masses were fixed as detailed in our previous study (Gao., 2019).
2.2 Light Microscopy (LM) Analyses
Spermatid masses were slightly centrifuged and fixed in Bouin’s solution for 24h at 4℃ and then embedded in paraffin. The paraffin was cut into 7µm serial sections. The sections were counterstained with hematoxylin and eosin and examined under a light microscope (Olympus BX51).
2.3 Transmission Electron Microscopy (TEM) Analyses
The spermatid masses were fixed in 2.5% glutaralde- hyde in cacodylate buffer (pH 7.4) at 4℃for 2h and then post-fixed for 1–2h in 1% osmium tetroxide. After dehy- drating in a graded ethanol series and acetone (30%, 50%, 70%, and 90%), cell masses were embedded in epoxy resin. Sections were cut using an ultramicrotome (LKB-α), stain- ed with uranyl acetate for 40min, and then counterstained with lead citrate (1min). A transmission electron micro- scope (JEM-1200EX) was employed to detect the morpho- logical characteristics of the sperm.
2.4 Cloning and Sequencing
Total RNA was extracted from the coelomic cells using Trizol reagent following the instruction of the manufac- turer (Takara, Tokyo, Japan). Thereafter, RNA quality was evaluated. The first-strand cDNA was obtained using the PrimerScriptTM RT reagent kit (Takara, Japan). To clone the intermediate fragments of,we analyzed theamino acid sequences downloaded from the National Cen- ter for Biotechnology Information (http://www.ncbi.nlm. nih.gov/, NCBI) and designed degenerate primers by us- ing an online tool (http://blocks.fhcrc.org/blocks/make_ blocks.html) Intermediate fragments were cloned under the following PCR conditions: 94℃ for 5min; 8 cycles of a touch-down program at 94℃ for 30s, 62℃ for 40s (0.5 ℃ reduction per cycle), and 72℃ for 40s; 30 cycles of 94 ℃ for 30s, 58℃ for 40s, and 72℃ for 40s; then 72℃ for 10min. The amplification products were disposed of fol- lowing the procedures described by Zhao(2017). Specific primers for 3’ and 5’ RACE were designed using Primer Premier 5.0 software (Premier Biosoft, CA, USA) on the basis of the intermediate chain of. RACE was performed using the SMARTerTMRACE cDNA Am- plification Kit (Clontech, CA, USA) in accordance with the manufacturer’s instructions (Primers see Table 1).

Table 1 Primers used in this study
2.5 Sequence and Phylogenetic Analysis
The information in NCBI (http://www.ncbi.nlm.nih.gov/)was used for sequence analysis. Subcellular localization was predicted on the website http://www.psort.org/. Pro- tein sequences were analyzed and aligned using the Vec- tor NT110 software (Invitrogen, California, USA). A phy- logenetic tree was constructed on Mega 5.4. The sequencesof KIF3A from human beings and different animals were used in protein alignment and phylogenetic analysis,in- cluding[XP_005105009.1],[AFP33455.1],[NP_001017604.2],[CAJ45482.1],[NP_001084268.1],[ELK03105.1],[XP_015973953.1],[XP_006820259.1],[EKC19840.1],[XP_ 013384616.1],[XP_013781360.1], and[NP_523934.1]. Sequences of KIF3B from hu-man beings and different animals were also employed in protein alignment and phylogenetic analysis, including[AEL16465.1],[NP_001093615.1],[NP_032470.3],[NP_001081489.1],[XP_005108306.1],[NP_ 001099999.1],[NP_004789.1],[AIN 36848.1], and[NP_001261726.1]. Protein do- mains and 3D structure were processed by PROSITE (http:// prosite.expasy.org/) and I-TASSER (http://zhanglab.ccmb. med.umich.edu/I-TASSER), respectively.
2.6 Real-Time Quantitative PCR
Specific primers were designed to analyze the expres- sion ofin different tissues. GAPDH served as a positive control (Su, 2010). The primers used are listed in Table 1.expression levels in different tissues were detected by qPCR as described by Zhao. (2017). The PCR process was 95℃ for 30s and 40 cycles at 95℃ for 20s, 60℃ for 20s, and 72℃ for 20s. All sam- ples were examined in quintuplicate on the sane plate (=5). Data processing and analysis were performed using SPSS 11.0.
2.7 Fluorescence in Situ Hybridization
Dual-color FISH was performed to identify the localiza- tion of/mRNA duringspermiogene- sis. The probes were designed by Primer 5.0 and synthe- sized by Invitrogen (Shanghai, China). Theantisense probe was a FITC-labeled 21bp-nucleotide sequence, and theantisense probe was a Cy3-labeled 24bp-nucleo- tide sequence (Primers are in Table 1).
Frozen sections of the experimental group and negative control group were incubated with probes (Gao., 2019). Then, the sections were observedconfocal laser-scan- ning microscopy (LSM710/780; Carl Zeiss, Germany).
2.8 Antibodies
Polyclonal rabbit anti-KIF3A and polyclonal rat anti- KIF3B were obtained as follows. The fragments in the tail domains of KIF3A (Ala-515 to Glu-687) and KIF3B (Ala- 503 to Arg-742) were cloned and ligated into the PEASY- Blunt E1 expression vector (TransGen, Beijing, China), transformed into Trans-T1 competent cells (Fig.1D), and sequenced by Beijing Genomics Institute (Beijing, China). Correct plasmids were extracted using the AxyPre Plas- mid Miniprep Kit (Axygen; Beijing, China) and transform- ed into Transetta (DE3) competent cells to produce the KIF3A-His6 and KIF3B-His6 fusion protein, induced by 1µmolL−1isopropylthio-β-d-galactoside (Fig.1A). The His- tag Protein Purification Kit was used to purify the fusion protein (Beyotime, Shanghai, China). Protein renaturation was performed similarly to our previous study (Gao., 2019) (Fig.1B). Finally, the recombination proteins were sent to Hangzhou Hua’an Biotechnology Co. Ltd. (China) for animal immunization. The specificity of antibodies was detected using Western blot (WB) as previously described (Zhao, 2017). The results agreed with the predicted molecular weights (Fig.1C).
The mouse anti-tubulin antibody was purchased from Be-yotime Biotechnology to detect the MT (Shanghai, Chi- na), and its specificity was analyzed by using WB (data not shown). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG and HRP-conjugated goat anti-rat IgG were purchased from Sangon Biotech (Shanghai, China). Alexa Fluor 488-labeled goat anti-mouse IgG (H+L), Alexa Fluor 488-labeled goat anti-rabbit IgG (H+L), Alexa Fluor 555- labeled goat anti-rabbit IgG (H+L), and Alexa Fluor Cy3- labeled goat anti-rat IgG (H+L) were purchased from Be- yotime Biotechnology. The Mito-Tracker Green FM was purchased from Invitrogen (California, USA).
2.9 Immunofluorescence (IF) Analyses
Frozen sections were cut and blocked as in our previ- ous work (Zhao., 2017). Then the frozen sections wereincubated with primary antibodies overnight (4℃) and washed with 1×PBS containing 0.1% Triton X-100 (0.1%PBST) in the concentrator (3 times, 15min per times). Af- ter incubation with secondary antibodies for 1 h at room temperature, the sections were washed six times with 0.1% PBST. Finally, the nuclei were stained with DAPI for 5min. The immunostained tissue sections were observed us- ing a confocal laser-scanning microscope (LSM710/780; Carl Zeiss, Germany). In the negative control, no primary antibody was added.

Fig.1 Preparation and validation of antibodies. (A) Expressions of recombinant proteins. The red boxes in Lines 1 and 4 show the expressions of recombinant Pe-KIF3A/3B, respectively. The other lines show the control groups. (B) Purification of recombinant proteins. Lines 3 and 4 show the purified recombination Pe-KIF3A, Lines 5 and 6 showed the control groups, Lines 9 and 10 show the purified recombination Pe-KIF3B. (C) Validation of antibodies. WB was employed to detect the specificity of two polyclonal antibodies. Only a single band appeared in each experimental group and coincided with the predicted molecular weight. (D) Immunogens of Pe-KIF3A/3B.
3 Results
3.1 Morphological Characteristics of Spermatogenic Cells in P. esculenta
By observing histological sections, we analyzed the mi-crostructures of spermatogenic cells of(Fig.2). According to the size, location, and shapes of the chroma- tin, germinal cells were divided into four groups: sperma- togonium, primary spermatocyte, secondary spermatocyte, and spermatid. Irregular-shaped spermatogonia were lo- cated at the near end of the testis with large cell bodies. Their nucleoli were close to the nuclear envelope (Fig.2A). The primary spermatocytes were located at the far-end of the testis with an ovoid-shaped nucleus (Fig.2B). At this stage, small particle-shaped chromatin was randomly dis- tributed in the nucleus. Secondary spermatocytes were most- ly located at the far end of the testis, with some floating spermatocytes in the coelomic fluid. Compared with pri- mary spermatocytes, secondary spermatocytes were small- er in size and more regular in shape. In addition, their nu- clei were approximately circular, and the chromatin was dispersed into small granules (Fig.2C). Finally, few sper- matids were located at the edge of the far-end of the testis, and most cells were free-floating in the coelomic fluid in the form of cell masses (Fig.2D).
The spermiogenesis and spermatid structure inwere characterized using TEM. In the early stage of spermiogenesis, the newly formed spermatids had round nuclei, the chromatin was not condensed (Figs.2E and 3A), and mitochondria were distributed around the nuclei (Fig.3A).Subsequently, visible proacrosomal vesicles formed by the Golgi apparatus were scattered in the cytoplasm. The ini- tial segment of the flagellum could be observed at this stage (Fig.3B). At the middle stage, the amount of cyto- plasm was reduced, as was the cell body size (Fig.2F). Chro-matin appeared in patches, and mitochondria began to moveto one cell pole. Some proacrosomal vesicles fused into short-flattened acrosomal vesicles (Fig.3C). In the late- stage spermatids, most of the cytoplasm was lost, the kar- yotheca appeared in irregular shapes, and the chromatin was distributed in the nucleus in the form of compact fine particles (Fig.2G). A mitochondria-rich region appeared. The flagellum and Golgi-centriolar complexes were clearly vi- sible (Fig.3D). The mature spermatid can be divided into two groups, spermatid with mature structure (stored in the coelomic fluid) and physically mature spermatid (stored in the nephridium) (Zhu., 2007). In the coelomic fluid, the mature spermatids were irregular in shape, the chroma- tin was highly concentrated, and the cytoplasm could be ob- served at one cell pole (Figs.2H and 2E). In the nephridium lumen, the sperm exhibited a substantial shape change; a cap-shaped acrosome appeared, and the nucleus exhibited a pear shape. A short midpiece and a slender flagellum witha typical ‘9+2’ structure could be observed (Figs.3I and 3F).
3.2 cDNA Sequence and Structural Analysis of Pe-kif3a and Pe-kif3b
The-(GenBanK number: KY411164) cDNA se- quence was 2500bp, with a 293bp 3’ UTR (untranslated re- gion), a 137bp 5’ UTR, and a 2070bp open reading frame (ORF). The polypeptide had a predicted molecular mass of 78.1kDa and a theoretical isoelectric point of 6.38.- KIF3A had a conserved central catalytic motor domain (8–341aa) containing eight ATP-binding sites (17-Arg, 96-Gly,99-Gly, 101-Gly, 102-Lys, 103-Thr, 104-Phe, and 246-Asp) and three MT-binding sites (294-Arg, 297-Lys, and 300- Arg). The neck linker (KNIKNKPRINEDPKDAL) could connect the head domain and the coiled–coil domain; it was located between residues 339 and 355. Moreover, we detected bipartite NLS motifs (KRHRRKKK, PKTGKKK) and bipartite site RRKLEQQKDMEEEEKRRV (Fig.4a).
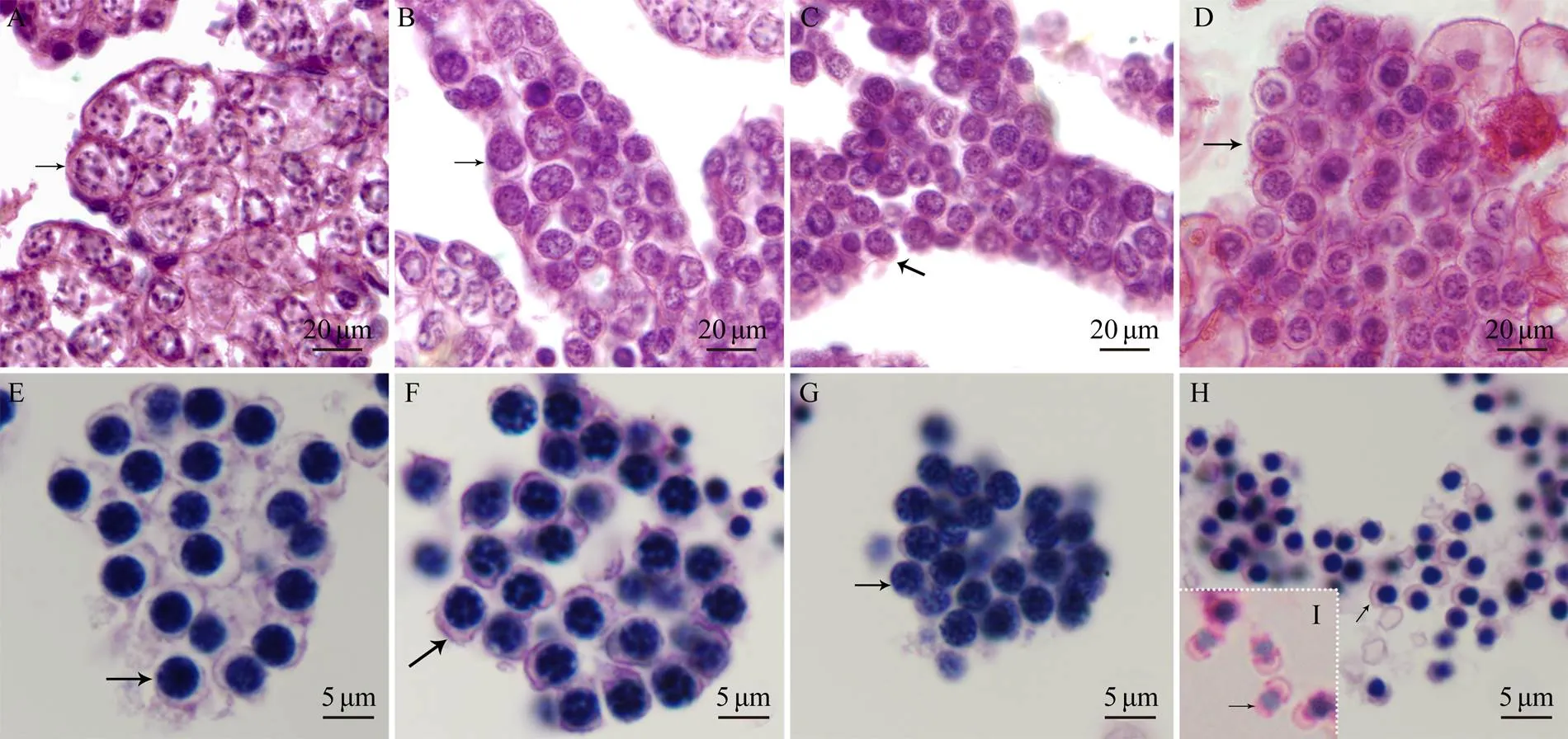
Fig.2 Microstructural characteristics of spermatogenesis in P. esculenta. Microstructures of spermatogenetic cells were analyzed by observing histological sections with HE staining: (A) spermatogonium, (B) primary spermatocyte, (C) secondary spermatocyte, (D) spermatid, (E) early spermatid, (F) middle spermatid, (G) late spermatid, (H) spermin coelomic fluid, and (I) sperm in nephridium lumen. The arrows indicate spermatids in the corresponding stage.

Fig.3 Ultrastructural characteristics of spermiogenesis in P. esculenta. TEM was employed to analyze the ultrastructures of spermiogenesis: (A) newly formed spermatid, (B) early spermatid, (C) middle spermatids, (D) late spermatids, (E) spermin coelomic fluid, and (F) sperm in nephridium lumen. N, nucleus; M, mitochondrion; F, flagellum; G, Golgi apparatus. The dotted circles indicate the proacrosomal vesicles. The triangle indicates the abandoned cytoplasm. The arrows (black and white) indicate the mitochondria.

Fig.4afull-length cDNA and amino acid sequence. The initiation codon (ATG) and termination codon (TGA) are set to yellow and blue, respectively. The red circles show the ATP bind sites, and the black boxes show the MT- binding sites. The blue line shows the neck linker.
Fig.4b Same as Fig.4a but forfull-length cDNA and amino acid sequence.
The full-lengthwas 3259 bp (GenBanK num- ber: KY411165), consisting of an 841-bp 3’ UTR, a 174bp5’ UTR, and a 2244bp ORF. The polypeptide had a pre- dicted molecular mass of 84.7kDa and a theoretical iso- electric point of 7.64. It also had ATP-binding sites (21-Arg,100-Gly, 103-Gly, 105-Gly, 106-Lys, 107-Thr, 108-Phe, and249-Asp) and MT-binding sites (297-Arg, 300-Lys, and 303- Arg) in the motor domain (11–344aa). The neck linker was located between residues 342 and 357.-KIF3B may be located in the nucleus, nucleolus, cytoplasm, and mito- chondria. In addition, we detected an NLS motif (PGKK KKKKRR) (Fig.4b).
The predicted secondary structure revealed that- KIF3A and-KIF3B could be divided into three do- mains (Fig.5A).-KIF3A was composed of a head at the N-terminal end, a helical stalk (342–583aa) and a tail (584–689aa) at the C-terminal end.-KIF3B was also composed of a head (1–344aa), a helical stalk (345–583aa), and a tail (584–747aa). The head was the catalytic core responsible for ATP-hydrolysis and MT binding. The 3D structural predictions for-KIF3A and-KIF3B weresimilar, including a globular head, a coiled-coil neck, and a globular tail (Fig.5B).
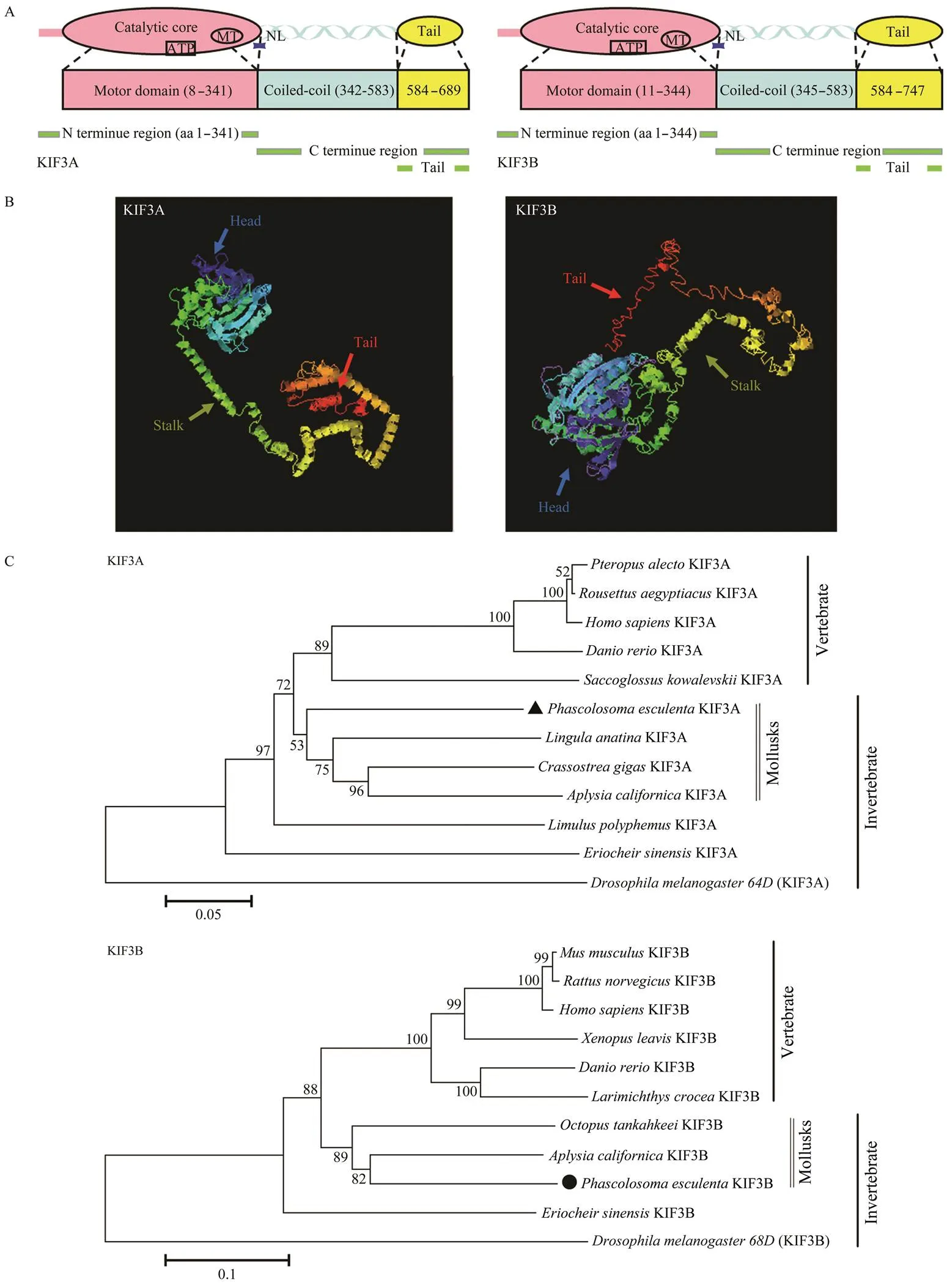
Fig.5 Structural prediction of Pe-KIF3A/3B proteins. A, Putative diagram of Pe-KIF3A/3B secondary structure; B, Putative diagram of Pe-KIF3A/3B tertiary structure; C, Phylogenetic analysis of Pe-KIF3A/3B. To construct the phylogenetic tree (NJ method), Mega 5.1 software was employed. Scale bar: 0.1 of the branch length value.
3.3 Sequence Alignment and Phylogenetic Analysis
The sequence of-KIF3A shared 77%, 57.3%, 66.6%, 71.8%, 69.2%, and 71.2% identity with the homologous proteins in,,,,,and, respectively.- KIF3A had high homology with sequences in other spe- cies, especially in its putative motor domain. In addition, the MT-binding sequence (YNEEVRDLL) and ATP-bind- ing sequence (AYGQTGTGKT, SSRSH, and LAGSE) (Fig.6A) were shared among species. The identities of KIF3B betweenand,,,,andwere 70.3%, 66.0%, 69.1%, 66.9%, and 73.5%, respectively. The se- quence of Pe-KIF3B also shared the MT-binding sequence (HLPYRDSKLTRLL) and ATP-binding sequence (VVV RCRP, NGTIFA, GQTGTGKT and DGENHIRVGKLNL VDLAGSERQ) with those of other species (Fig.6B).- KIF3A/3B showed low conservation in non-motor do- mains.
We generated a phylogenetic tree by the neighbor- joining (NJ) method to evaluate-KIF3A and-KIF3B in an evolutionary framework.KIF3A shared the high-est similarity with homologs in the mollusksand, and-KIF3B shared the highest similarity with homologs in the mollusksand(Fig.5C).
3.4 Relative Abundance of Pe-kif3a/3b mRNA
The purified spermatid mass, intestine, nephridium, re- tractor muscle, and body wall were obtained from adult worms and quantified by real-time quantitative PCR to examine the relative abundances ofandtran- scripts in.mRNAs were detected in all tissues assayed (Fig.7). The sperm masses had the highest abundances of, suggesting that these loci play crucial roles in, especially in sper- miogenesis.
3.5 Dynamic Expression Patterns of Pe-kif3a/3b mRNA During Spermiogenesis
FISH was employed to precisely determine the tempo- ral and spatial expression patterns ofduring spermiogenesis in. Negative controls with sense probes were used to detect the specificity of fluorescence signals (data not shown). The expression patterns of thetwo genes were similar. In the early stage of spermatids,andexhibited low expression levels, andsignals were sporadically distributed in the cytoplasm (Figs.8A1–A4). In the middle stage of spermatids,andexhibited remarkably elevated expres- sion levels distributed evenly throughout the cytoplasm(Figs.8B1–B4). In the late stage of spermatids, the expres- sion patterns ofandwere restricted, withdense signals at one cell (Figs.8C1–C4) pole. In mature sperm,andsignals were intense in the subacro- somal space and tail midpiece (Figs.8D1–D4).

Fig.6A Multiple sequence alignment of KIF3A homologous proteins. Theblue boxes indicate the putative ATP-binding sites (AYGQTGTGKT, SSRSH, and LAGSE). The red boxes indicate the putative microtubule binding sites (YNEE VRDLL).
Fig.6B Multiple sequence alignment of KIF3B homologous proteins. Theblue boxes indicate the putative ATP-binding sites (VVVRCRP, NGTIFA, GQTGTGKT, and DGENHIRVGKLNLVDLAGSERQ). The red boxes indicate the putative microtubule binding sites (HLPYRDSKLTRLL).

Fig.7 Relative abundance of Pe-kif3a/3b mRNA in different tissues. Relative abundance levels of Pe-kif3a (black bars) and Pe-kif3b (gray bars) in different tissues were detected by qPCR. Pe-kif3a and Pe-kif3b show the highest expression levels in spermatid mass. S,spermatid mass; I, intestine; N, nephridium; Rm, retractor muscle; Bw, body wall. All samples were ex- amined in quintuplicate on the same plate (n=5).
3.6 Co-Localization of Pe-KIF3A/3B with MTs During P. esculenta Spermiogenesis
Using immunofluorescence, we analyzed the protein expression characteristics of-KIF3A,-KIF3B, and MTs (Figs.9, 10). In early spermatids, each of the three proteins exhibited weak expression in the cytoplasm, and-KIF3A/3B co-localized with MTs (Figs.9, 10A1–A4). In intermediate spermatids, the signals of MTs and- KIF3A/3B were enhanced and co-localized in the cyto- plasm (Figs.9, 10B1–B4). In late spermatids, the MTs sig- nals were dense and were tightly packed in the nucleus, whereas-KIF3A/3B were detected in the karyotheca at one pole (Figs.9, 10C1–C4). In the sperm, the expression levels of both proteins were remarkably reduced.-KIF3A/ 3B signals were detected in the midpiece and flagellum, and the MT signals were co-localized with KIF3A/B in the flagellum (Figs.9, 10D1–D4).
3.7 Pe-KIF3A/3B Mediated the Migration of Mito- chondria During P. esculenta Spermiogenesis
Immunofluorescence was performed to detect whether KIF3A/3B serve mitochondria-related functions duringspermiogenesis. Notably,-KIF3A andKIF3B co-localized with mitochondria. The co-localized signals could be observed during spermiogenesis (Figs.11, 12). In early spermatids, few mitochondria and-KIF3A/3B were evenly distributed in the cytoplasm. (Figs.11, 12A1–A4). In the middle spermatids, the co-localization increased (Figs.11, 12B1–B4). The migration of mitochondria was initially detected in the middle spermatids, and the signals were located in the postnuclear cytoplasm (Figs.11, 12 C1– C4). In addition, co-localization was detected in the mid- piece of the sperm tail (Figs.11, 12D1–D4).
3.8 Co-Localization of Pe-KIF3A with Pe-KIF3B During Spermiogenesis
Immunofluorescence showed thatKIF3A co-localiz- ed with-KIF3B at all stages ofspermio- genesis (Fig.13). In the early and middle stages,-KIF3A and-KIF3B were both localized around the nucleus. In the late spermatids, both proteins were localized along one side of the nucleus. In the mature sperm, the co-localized signals were observed at the midpiece of the tails. The co-localization of-KIF3A and-KIF3B implied the heterodimerization of the two proteins as well as their po- tential functions in spermiogenesis. These results provide a basis for a model of the distribution of KIF3A and KIF3B during spermiogenesis in(Fig.14a).
4 Discussion
In this study, we observed several noteworthy morpho- logical changes duringspermiogenesis. 1) We observed a decrease in cell size. In our previous study, we found that the cytoplasm forms a pseudopod-like protu- berance in middle-stage spermatids, which is later lost by exocytosis, leading to a decrease in cell size (Long., 2015; Gao., 2019). In mammals, exocytosis is regu- lated by MT-dependent IFTKinesin II (Finetti., 2009). However, whether-KIF3A/3B functions in a si- milar manner duringspermiogenesis is un- clear. 2) With respect to sperm head deformation, we ob- served acrosome biogenesis and nuclear reshaping. MTs likely facilitate sperm head deformation in mice (O’ Don- nell and O’ Bryan, 2014). We found that MTs surrounded the nuclei during these processes (Figs.9, 10). 3) The en- flagellation and migration of mitochondria were observed. Overall, several significant morphological changes were observed during spermiogenesis in,and these changes were associated with MTs and Kinesin3A/3B. These findings are discussed in detail below.
4.1 Bioinformatics and Expression Analyses of Pe-kif3a/3b
KIF3A and KIF3B can be divided into the N-terminal motor domain, coiled-coil domain, and C-terminal tail do- main (De Cuevas., 1992). Using online prediction tools, we found that Pe-KIF3A/3B had these structures, in-cluding highly conserved motor domains containing AT- Pase and MT-binding sites. The tail domain can interact with other subunits of holoenzymes and cargoes (Miki., 2005; Ma., 2017). Coiled-coil domains act as bridges for interactions between KIF3A and KIF3B (Yamazaki., 1994; Yamazaki., 1996; Chana., 2005; Gu- zik-Lendrum., 2015; Mu., 2019), enabling he- tero-dimerization (Rashid., 1995). In the present study, the coiled-coil domains in-KIF3A/3B were relatively conserved, suggesting that-KIF3A/3B is also hetero- dimerized in.
mRNA expression patterns are usually associated with functions of the proteins.mRNAs are found in various tissues and organs among species (Henson., 1997; Wang., 2010; Hu., 2012; Lu., 2014; Zhao., 2017). In our study,-mRNAs were detected in all tissues assayed (Fig.7). The highest tran- script abundances were found in sperm masses, suggest- ing their crucial roles in,especially in sper- miogenesis. Using FISH, we observedsignals at all stages of spermiogenesis but at low levels in the new- ly formed spermatid (Fig.8). In middle-stage spermatid,-expression levels reached a peak, and various morphological changes were observed by TEM (as shown in Fig.2), including chromatin condensation, enflagella- tion, mitochondria migration, and increased proacrosomalvesicles (Long., 2015). These results implied that the two genes might be involved in acrosome biogenesis, nu- clear deformation, and tail formation. In mature sperm, the signals were highly condensed and co-localized in the sub- acrosomal space and midpiece with high intensity. In ge- neral,mRNA is minimally or not expressed in ma-ture sperm (Miller., 1999a; Wang., 2010; Dang., 2012; Hu., 2012). Considering thatcould be detected in early embryos (Marszalek., 1999), we predict that the residual transcripts may enter eggs and thereby contribute toembryonic development.

Fig.8 Dynamic expression pattern of Pe-kif3a/3b during spermiogenesis. FISH was employed to analyze the expression pattern of Pe-kif3a (green) and Pe-kif3b (Red). DAPI shows the nuclei (blue). A1–A4 show the early spermatids.Pe- kif3a/3b were weakly expressed in the perinuclear cytoplasm. B1–B4 show the middle spermatids. The expression level of Pe-kif3a/3b reached a peak. C1–C4 show the late spermatids. The white arrow shows the signals, which were con- centrated and located at one side of the cell. D1–D4 show the mature sperm. The signals could be detected in the mid- piece and subacrosomal space. The scale bar is 10μm.

Fig.9 Localization of Pe-KIF3A and tubulin during P. esculenta spermiogenesis. IF assay shows the localization of Pe- KIF3A and tubulin. The tubulin signals (green) were co-localized with Pe-KIF3A (red) during all stages of spermiogene- sis. DAPI (blue) shows the nucleus. The arrows and dotted circles show the signals. A1–A4 show the early spermatids. Pe-KIF3A co-localized with tubulin in the perinuclear cytoplasm at a low level. B1–B4 show the middle spermatids. C1–C4 show the late spermatids. Co-localization of Pe-KIF3A and tubulin could be observed in the cytoplasm near the nuclear membrane. D1–D4 show the mature sperm. Signals co-localized at the midpiece. The scale bar is 10μm.

Fig.10 Localization of Pe-KIF3B and tubulin during P. esculenta spermiogenesis. IF assay shows the localization of Pe- KIF3B (red) and tubulin (green). DAPI (blue) shows the nucleus. The arrows and dotted circles show the signals. A1–A4show the early spermatids. B1–B4 show the middle spermatids. C1–C4 show the late spermatids. D1–D4 show the mature sperm. The scale bar is 10μm. The co-localization of Pe-KIF3B and tubulin shares the same pattern as Pe-KIF3A and tu- bulin.

Fig.11 Localization of Pe-KIF3A and Mito-tracker during P. esculenta spermiogenesis. IF was employed to analyze the co-localization of Pe-KIF3A and mitochondria. DAPI (blue) shows the nucleus. Mito-tracker signals (green) were co-lo- calized with Pe-KIF3A (red) during all stages. A1–A4 show the early spermatids. Pe-KIF3A co-localized with mitochon- dria in the perinuclear cytoplasm. B1–B4 show the middle spermatids. The co-localized signals were detected in the cy- toplasm. C1–C4 show the late spermatids. Co-localization of Pe-KIF3A and mitochondria could be observed on one side of the spermatid. D1–D4 show the mature sperm. Notably, they were co-localized at the midpiece. The scale bar is 10μm.

Fig.12 Localization of Pe-KIF3B and Mito-tracker during P. esculenta spermiogenesis. DAPI (blue) shows the nucleus. Mito-tracker signals (green) were co-localized with Pe-KIF3B (red). A1–A4 show the early spermatids. B1–B4 show the middle spermatids. C1–C4 show the late spermatids. D1–D4 show the mature sperm. The scale bar is 10μm.

Fig.13 Co-localization of Pe-KIF3A with Pe-KIF3B during P. esculenta spermiogenesis. IF assay shows the co-localization of Pe-KIF3A (green) and Pe-KIF3B (red). A1–A4 show the early spermatids.B1–B4 show the middle spermatids. C1–C4 show the late spermatids. D1–D4 show the mature sperm. Pe-KIF3A always co-localized with Pe-KIF3B. Co-local- ization indicates the heterodimerization of the two proteins. The scale bar is 10μm.

Fig.14 A, Model of the dynamic distribution of Pe-KIF3A/3B during spermiogenesis in P. esculenta. M, mitochondria; N, nucleus; F, flagellum. (a), in the early stage, Pe-KIF3A/3B were sporadically distributed in the cytoplasm; (b), in the middle stage, Pe-KIF3A/3B exhibited remarkably elevated expression levels; (c), in the late stage, the expression patterns of Pe- KIF3A/3B were restricted; (d), in mature sperm, Pe-KIF3A/3B signals were intense in the tail midpiece. The scale bar is 10μm. B, Model of Pe-KIF3A/3B putative functions in P. esculenta spermiogenesis. Based on our results, we suggested that (a) Pe-KIF3A/3B participate in enflagellation and flagellar maintenance through IFT; (b) Pe-KIF3A/3B may transport the acrosomal vesicles from the Golgi apparatus during acrosome biogenesis; (c) Pe-KIF3A/3B might participate in sperm head remodeling; (d) in addition, the two proteins are involved in mediating mitochondrial behavior during midpiece for- mation.
4.2 Pe-KIF3A/B and Sperm Head Remodeling
MTs are highly dynamic filaments with pivotal roles in many fundamental cellular processes, including division, motility, intracellular transport, and cell shaping (Jordan and Wilson, 2004; Conde and Caceres, 2009; Helmke.,2013). During spermiogenesis, MTs are involved in sperm head remodeling, including nuclear deformation and acro- some biogenesis (Fawcett., 1971; Moreno., 2006; Zhao., 2017). In this study, MT signals could be de- tected inspermatids at all stages (Figs.9 and 10).
4.2.1 Nucleus reshaping
MTs are essential for sperm head remodeling (Russell., 1991; Mendoza-Lujambio., 2002; Lehti andSironen, 2016; Dunleavy., 2017). Perinuclear MTs can interact with the nuclear envelope and karyoskeleton, thereby deforming spermatid nuclei by mechanical force (MacKinnon and Abraham, 1972; Rattner and Brinkley, 1972). As MT-dependent proteins, kinesins contribute to this process. In mice, KIF5C and KIFC1 interact with the manchette and promote nuclear deformation (Navolanic and Sperry, 2000; Kotaja., 2004; Saade., 2007). The depletion of KIF3A leads to abnormalities in the spermhead (Lehti., 2013). In addition to mammals, the functions of KIF3A/3B in nuclear deformation have been evaluated in many species, including,, and(Zhao., 2017; Mu., 2019; Wang., 2019). Our immuno- fluorescence analysis showed thatKIF3A/3B co-loca- lized with MTs throughout spermiogenesis, and their ex- pression levels were positively correlated with the degree of nuclear deformation (Figs.9 and 10). Notably,-KIF3A/3B signals reached a peak and were co-localized with peri- nuclear MTs in late-stage spermatids, along with substan- tial deformations, including chromatin condensation and nu- clear shrinkage. Therefore, our results suggest that-KIF3A/ 3B and MTs participate in the nuclear reshaping ofsperm.
4.2.2-KIF3A/B participating in the loss of cytoplasm
Cytoplasm abandonment decreases spermatid size. In, the pseudopodia-like cytoplasmic protrusionswere associated with this process. With the decline of pro-trusions in late spermatids, redundant cytoplasmic compo- nents and unnecessary organelles would be removed by autophagy and exocytosis. Gao(2019) reported that MTs and KIFC1 are involved in the generation and main- tenance of pseudopodia-like cytoplasmic protrusions inspermatids, indicating that cytoplasm removal relies on MTs and motor proteins.
Recent research has shown that KIF3A/3B play roles in the removal of redundant cytoplasm. As the core organelle in autophagy, lysosomes could be transported by KIF3A/ 3B (Brown., 2005). As a cargo of KIF3A/3B, IFT20 directs autophagy (Pampliega., 2013). Finally, Kine- sin II could regulate exocytosis (Finetti., 2009). With respect to the sperm, Zhang(2019) reported that IFT20 participates in the removal of cytoplasmic compo- nents by autophagy. Basing on the co-localization of MTs and-KIF3A/3B observed in our study, we suggest that-KIF3A/3B participates in cytoplasm abandonment du- ring spermiogenesis in.
4.2.3 Acrosome biogenesis
The acrosome is an important organelle for fertilization and is formed by Golgi-secreted proacrosomal vesicles (No- lan and Hammerstedt, 1997). The proacrosomal vesicles are transported to the acroplaxome and fused to form an acrosome vesicle, finally resulting in the acrosome. Dur- ing acrosome biogenesis, Kinesin II transports these vesi- cles along MTs (Moreno., 2006; Abraham., 2011). In, KIF3A and tubulin are co-localized in the lamellar complex, indicating their role in acrosome for- mation (Zhao., 2017). In the crustacean, KIF3A and KIF3B play crucial roles in acrosome bioge- nesis (Yu., 2009; Lu., 2014). In mammals, Ki- nesin II participates in acrosome biogenesis by transport- ing the Golgi apparatus, endoplasmic reticulum, and ly- sosomes (Stauber., 2006; Brown., 2010). We observed the subcellular localization of-KIF3A/3B and tubulin by immunofluorescence, consistent with the TEM results for the localization of proacrosomal vesicles. This observation implied that KIF3A/3B might transport the pro- acrosomal vesicle along MTs to form the acrosome.
4.3 Pe-KIF3A/3B Facilitating Sperm Tail Formation
Tails ofsperm can be divided into the end- piece and the midpiece. Enflagellation is initiated in early spermatids. As is typical of ciliogenesis, enflagellation oc- curs in an IFT-dependent manner (Dunleavy., 2019). Several kinesins participate in sperm tail formation, in- cluding KIFC1, KIFC5, KIF17b, KLC3, and KIF3AB (Ce- sario and Bartles, 1994; Navolanic and Sperry, 2000; Yangand Sperry, 2003), with functions in IFT and mitochon- drial transport. We focus on the roles of KIF3A/3B in tail formation in, especially in enflagellation and midpiece organization.
4.3.1-KIF3A/3B facilitating enflagellation
The elongation of the axoneme is the first step in en- flagellation, followed by the transport of other materials along axonemal MTs to assemble the flagellum. Consi- dering the dynamic instability of MTs, the material-traf- ficking cycles are needed to maintain the flagellum. Thus IFT is essential for sperm tail development and mainte- nance (Kozminski., 1993; Cole., 1998; Rosenbaumand Witman, 2002; Scholey, 2012). As an important an- terograde motor in IFT, Kinesin II controls enflagellation (Prevo., 2017). By the transport of the IFT-B com- plex, kinesin-2 and KIF3A/3B are responsible for ciliary assembly (Prevo., 2017; Funabashi., 2018).Re- cent research on the trafficking of the IFT38–IFT52–IFT57– IFT88 complex by KIF3A/3B supports this conclusion (Fu- nabashi., 2018). In addition, inmice, ciliary morphogenesis exhibits defects (Nonaka., 1998; Mars- zalek., 1999), and the depletion of KIF3A affects the elongation of the axoneme (Lehti., 2013). Moreover, IFT20, an essential subunit for enflagellation, moves in a KIF3A3/B-dependent manner (Zhang., 2019).In ear- ly spermatids, the flagellum is difficult to recognize, and the co-localized signals of KIF3A/3B and MTs are distri- buted evenly in the cytoplasm. With the elongation of the flagellum, the MT signals become dense in the postnu- clear cytoplasm, where the flagellum forms. Axonemal MTs can also be observed. Thus, the co-localization of KIF3A/ 3B and MTs (especially axonemal MTs) supports the rolesof-KIF3A/3B in sperm enflagellation. Notably,- KIF3A/3B and MTs co-localized in the flagellum of ma- ture sperm, suggesting that-KIF3A/3B participate in the maintenance of the flagellum. Overall,-KIF3A/3B promote flagellum formation during spermiogenesis and facilitate the maintenance of the flagellumIFT.
4.3.2-KIF3A/3B mediating mitochondrial behaviors during midpiece formation
The midpiece ofconsists of two centrioles and a helical mitochondrial sheath (Zhu., 2007). TEM showed the migration and fusion of mitochondria during spermiogenesis, which were initially evenly distributed in the cytoplasm and eventually fused into the midpiece (Fig.3). Immunofluorescence showed a similar distribution of mi- tochondria, andKIF3A/3B co-localized with mitochon- dria at all time points (Figs.11 and 12). These results in- dicate that-KIF3A/3B participate in midpiece formation in. Some studies have suggested that KIF3A/ 3B are involved in mitochondrial trafficking (Zhao., 2017; Wang., 2019), and the co-localization of- KIF3A/3B and MTs supports this hypothesis. Interesting- ly, heterotrimeric kinesin-2 could bind to dynactin (Muel- ler., 2005; Berezuk and Schroer, 2007), which can link dynein to the mitochondrial membrane surface and regulate mitochondrial transport (Deacon., 2003; Dre- rup., 2017). This result supports the hypothesis from a different perspective. However, compared with Kinesin- 1 and cytoplasmic dynein (Melkov and Abdu, 2017), evi- dence on the role of KIF3A/3B in mitochondrial transport is lacking. Notably, KIF3A/3B and dynein can bind to the same cargoes in bidirectional IFT (Andreasson., 2015). Thus, we postulated that in addition to the direct transport of mitochondria, KIF3A/3B might influence mitochondrial behaviors by mediating the functions of related proteins, such as dynein. In conclusion, we proposed that-KIF3A/3B influence mitochondrial behaviors during midpiece for-mation by direct effects on transport and indirect effects on other functions.
4.4 Heterodimeric KIF3AB Participating in Spermiogenesis of P. esculenta
Although KIF3A/3B could act in the monomeric form, our results suggest that the heterodimeric KIF3AB has cri- tical functions in. Compared with monomeric motors, dimeric motors have several advantages. First, in the ‘step-by-step’ model, two motor domains improve the coordination (Clancy., 2011; Mickolajczyk and Han- cock, 2017). Second, dimeric motors are efficient transpor-ters, especially under high-load conditions. Finally, dimers could work effectively at low concentrations (Schimert., 2019). In mammals, KIF3A and KIF3B could form heterodimeric KIF3AB in a preferential and spontaneous way (Chana., 2005). For example, the IFT-B complex could be transported by heterotrimeric kinesin-2. By con- trast, other than the KIF3ABheterodimer, the monomeric KIF3A/3B and dimers of KIF3A/3B could not completethese normal functions. Our immunofluorescence analysis showed thatKIF3A co-localized with-KIF3B at all stages of spermiogenesis, indicating the direct interaction between the two proteins. These findings suggest that- KIF3A/3B participate inspermiogenesis ofin the form of-KIF3AB.
5 Conclusions
This is the first study of KIF3A/3B proteins in. We provide a detailed description of spermiogene- sis in the species through HE staining and TEM. We de- tected the mRNA expression patterns of, which are ubiquitously expressed with particularly high levels in sperm masses. Immunofluorescence showed the consistent co-localization of MTs and-KIF3A/3B, indicating their potential functions in sperm head remodeling and tail fo- rmation. The co-localization of-KIF3A/3B and mito-chondria suggests that they function in midpiece forma- tion because the functions of KIF3A/3B are conservative. Therefore, a creditable model of protein functions was pro- posed (Fig.14b). In addition, our findings suggest that he- terodimeric-KIF3AB participates in sperm head re- modeling, including nuclear reshaping, cytoplasm aban- donment, and acrosome biogenesis.-KIF3AB also has potential functions in sperm tail formation by facilitatingsperm enflagellation and mediating mitochondrial beha- vior during midpiece formation.
Acknowledgements
The authors are grateful to all members of the Fish His- tology Laboratory in Ningbo University for providing di- rect assistance and constructive discussions for this re- search. This project was supported by the Ningbo Science and Technology Plan Projects (Nos. 2019B10016, 2016C 10004), the Major Science and Technology Projects in Zhejiang Province (No. 2011C12013), the Natural ScienceFoundation of Zhejiang Province (No. LY18C190007), the National Natural Science Foundation of China (No. 312 72642), theCollaborative Innovation Center for Marine High-efficiency and Healthy Aquaculture of Zhejiang Pro-vince, and K. C. Wong Magna Fund in Ningbo University.
Amaral, A., Lourenço, B., Marques, M., and Ramalhosantos, J., 2013. Mitochondria functionality and sperm quality., 146 (5): 163-174.
Andreasson, J. O., Shastry, S., Hancock, W. O., and Block, S. M., 2015.The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load., 25 (9): 1166-1175.
Berezuk, M. A., and Schroer, T. A., 2007. Dynactin enhances the processivity of kinesin-2., 8 (2): 124-129.
Brown, C. L., Maier, K. C., Stauber, T., Ginkel, L. M., Wordeman,L., Vernos, I.,., 2005. Kinesin-2 is a motor for late endo- somes and lysosomes., 6 (12): 1114-1124.
Cesario, M. M., and Bartles, J. R., 1994. Compartmentalization, processing and redistribution of the plasma membrane protein CE9 on rodent spermatozoa., 107 (4): 561-570.
Chana, M. S., Tripet, B. P., Mant, C. T., and Hodges, R., 2005. Stability and specificity of heterodimer formation for the coiled- coil neck regions of the motor proteins KIF3A and KIF3B: The role of unstructured oppositely charged regions.,65 (2): 209-220.
Clancy, B. E., Behnke-Parks, W. M., Andreasson, J. O., Rosen- feld, S. S., and Block, S. M., 2011. A universal pathway for ki- nesin stepping., 18 (9): 1020-1027.
Cole, D. G., Diener, D. R., Himelblau, A. L., Beech, P. L., Fuster, J.C., and Rosenbaum, J. L., 1998. Chlamydomonas kinesin-II-de- pendent intraflagellar transport (IFT): IFT particles contain pro- teins required for ciliary assembly insensory neurons., 141 (4): 993-1008.
Conde, C., and Caceres, A., 2009. Microtubule assembly, organi- zation and dynamics in axons and dendrites., 10 (5):319-332.
Dang, R., Zhu, J. Q., Tan, F. Q., Wang, W., Zhou, H., and Yang, W. X., 2012. Molecular characterization of a KIF3B-like kine- sin gene in the testis of(Cephalopoda, Octopus)., 39 (5):5589-5598.
Deacon, S. W., Serpinskaya, A. S., Vaughan, P. S., Lopez-Fanar- raga, M., Vernos, I., Vaughan, K. T.,., 2003. Dynactin is re- quired for bidirectional organelle transport., 160 (3): 297-301.
DeCuevas, M., Tao, T., and Goldstein, L. S. B., 1992. Evidence that the stalk ofkinesin heavy chain is an a-heli- cal coiled coil., 116: 957-965.
Drerup, C. M., Herbert, A. L., Monk, K. R., and Nechiporuk, A. V., 2017. Regulation of mitochondria-dynactin interaction and mitochondrial retrograde transport in axons., 6: e22234.
Dunleavy, J. E. M., O’Bryan, M. K., Stanton, P. G., and O’Donnell, L., 2019. The cytoskeleton in spermatogenesis., 157 (2): 53-72.
Dunleavy, J. E., Okuda, H., O’connor, A. E., Merriner, D. J.,O’Donnell, L., Jamsai, D.,., 2017. Katanin-like 2 (KATNAL2) functions in multiple aspects of haploid male germ cell deve- lopment in the mouse., 13 (11): e1007078.
Fan, J., and Beck, K. A., 2004. A role for the spectrin superfa- mily member Syne-1 and kinesin II in cytokinesis., 117 (4): 619-629.
Fawcett, D. W., Anderson, W. A., and Phillips, D. M., 1971. Mor- phogenetic factors influencing the shape of the sperm head., 26 (2):220-251.
Finetti, F., Paccani, S. R., Riparbelli, M. G., Giacomello, E., Peri- netti, G., Pazour, G. J.,., 2009. Intraflagellar transport is required for polarized recycling of the TCR/CD3 complex to the immune synapse., 11 (11): 1332-1339.
Funabashi, T., Katoh, Y., Okazaki, M., Sugawa, M., and Nakayama, K., 2018. Interaction of heterotrimeric kinesin-II with IFT-B-connecting tetramer is crucial for ciliogenesis., 217 (8):2867-2876.
Gao, X. M., Mu, D. L., Hou, C. C., Zhu, J. Q., Jin, S., and Wang, C. L., 2019. Expression and putative functions of KIFC1 for nuclear reshaping and midpiece formation during spermioge- nesis of., 683:169-183.
Gilbert, S. P., Guzik-Lendrum, S., and Rayment, I., 2018. Kine- sin-2 motors: Kinetics and biophysics., 293 (12): 4510-4518.
Guzik-Lendrum, S., Rank, K. C., Bensel, B. M., Taylor, K. C., Rayment, I., and Gilbert, S. P., 2015. Kinesin-2 KIF3AC and KIF3AB can drive long-range transport along microtubules., 109 (7): 1472-1482.
Hall, E. S., Eveleth, J., Jiang, C., Redenbach, D. M., and Boekel-heide, K., 1992. Distribution of the microtubule-dependentmotors cytoplasmic dynein and kinesin in rat testis., 46 (5):817-828.
Haraguchi, K., Hayashi, T., Jimbo, T., Yamamoto, T., and Akiyama, T., 2006. Role of the kinesin-2 family protein, KIF3, during mitosis., 281 (7): 4094-4099.
Helmke, K. J., Heald, R., and Wilbur, J. D., 2013. Interplay be- tween spindle architecture and function., 306: 83-125.
Henson, J. H., Cole, D. G., Roesener, C. D., Capuano, S., Men- dola, R. J., and Scholey, J. M., 1997. The heterotrimeric mo- tor protein kinesin-II localizes to the midpiece and flagellum of sea urchin and sand dollar sperm., 38 (1): 29-37.
Hirokawa, N., and Noda, Y., 2008. Intracellular transport and ki-nesin superfamily proteins, KIFs: Structure, function, and dyna- mics., 88 (3):1089-1118.
Hu, J. R., Xu, N., Tan, F. Q., Wang, D. H., Liu, M., and Yang, W. X., 2012. Molecular characterization of a KIF3A-like kinesin gene in the testis of the Chinese fire-bellied newt., 39 (4): 4207-4214.
Jordan, M. A., and Wilson, L., 2004. Microtubules as a target for anticancer drugs., 4 (4):253-265.
Kierszenbaum, A. L., Rivkin, E., and Tres, L. L., 2011. Cyto- skeletal track selection during cargo transport in spermatids is relevant to male fertility., 1 (3):221-230.
Kodani, A., Salomé, S. M., Seol, A., Garciaverdugo, J. M., and Reiter, J. F., 2013. Kif3a interacts with Dynactin subunit p150Gluedto organize centriole subdistal appendages., 32 (4):597-607.
Kotaja, N., De-Cesare, D., Macho, B., Monaco, L., Brancorsini, S., Goossens, E.,., 2004. Abnormal sperm in mice with tar- geted deletion of the act (activator of cAMP-responsive element modulator in testis) gene., 101 (29): 10620-10625.
Kozminski, K. G., Johnson, K. A., Forscher, P., and Rosenbaum, J. L., 1993. A motility in the eukaryotic flagellum unrelated to flagellar beating., 90 (12): 5519-5523.
Lehti, M. S., and Sironen, A., 2016. Formation and function of the manchette and flagellum during spermatogenesis., 151 (4): R43-R54.
Lehti, M. S., Kotaja, N., and Sironen, A., 2013. KIF3A is essen- tial for sperm tail formation and manchette function., 377 (1-2): 44-55.
Li, F. L., 1992. A checklist of sipuncula from the China coasts., 22 (2): 72-88.
Long, L. L., Sheng, Z., and Zhu, J. Q., 2015. Ultrastructural ob- servations on spermiogenesis in the peanut worm,(Sipuncula:)., 19 (3): 1-9.
Lu, Y., Wang, Q., Wang, D. H., Zhou, H., Hu, Y. J., and Yang, W. X., 2014. Functional analysis of KIF3A and KIF3B during spermiogenesis of Chinese mitten crab., 9 (5): e97645.
Ma, D. D., Wang, D. H., and Yang, W. X., 2017. Kinesins in spermatogenesis., 96 (2): 267-276.
MacKinnon, E. A., and Abraham, J. P., 1972. The manchette in stage 14 rat spermatids: A possible structural relationship with the redundant nuclear envelope., 124 (1):1-11.
Marszalek, J. R., and Goldstein, L. S., 2000. Understanding the functions of kinesin-II., 1496 (1):142-150.
Marszalek, J. R., Liu, X., Roberts, E. A., Chui, D., Marth, J. D., Williams, D. S.,., 2000. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors., 102 (2):175-187.
Marszalek, J. R., Ruiz-Lozano, P., Roberts, E., Chien, K. R., and Goldstein, L. S., 1999. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II., 96 (9): 5043-5048.
Melkov, A., and Abdu, U., 2017. Regulation of long-distance tran- sport of mitochondria along microtubules., 75 (2): 163-176.
Mendoza-Lujambio, I., Burfeind, P., Dixkens, C., Meinhardt, A., Hoyer-Fender, S., Engel, W.,., 2002. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh)mutant mouse., 11 (14): 1647-1658.
Mickolajczyk, K. J., and Hancock, W. O., 2017. Kinesin processi- vity is determined by a kinetic race from a vulnerable one-head- bound state., 112 (12):2615-2623.
Miki, H., Okada, Y., and Hirokawa, N., 2005. Analysis of the ki- nesin superfamily: Insights into structure and function., 15 (9):467-476.
Miller, D., Briggs, D., Snowden, H., Hamlington, J., Rollinson, S.,Lilford, R.,., 1999a. A complex population of RNAs exists in human ejaculate spermatozoa: Implications for understand- ing molecular aspects of spermiogenesis., 237 (2): 385-392.
Miller, M. G., Mulholl, D. J., and Vogl, A. W., 1999b. Rat testis motor proteins associated with spermatid translocation (dynein) and spermatid flagella (kinesin-II)., 60 (4): 1047-1056.
Moreno, R. D., Palomino, J., and Schatten, G., 2006. Assembly of spermatid acrosome depends on microtubule organization dur- ing mammalian spermiogenesis., 293 (1): 218-227.
Mu, D. L., Du, C., Fu, S. Y., Wang, J. Q., Hou, C. C., Tang, D. J.,., 2019. Molecular characterization, tissue distribution and localization ofKif3a and Kif3b and ex- pression analysis of their genes during spermiogenesis., 18 (6): 1451-1469.
Mueller, J., Perrone, C. A., Bower, R., Cole, D. G., and Porter, M. E., 2005. The FLA3 KAP subunit is required for localiza- tion of kinesin-2 to the site of flagellar assembly and proces- sive anterograde intraflagellar transport., 16 (3): 1341-1354.
Navolanic, P. M., and Sperry, A. O., 2000. Identification of iso- forms of a mitotic motor in mammalian spermatogenesis., 62 (5):1360-1369.
Nolan, J. P., and Hammerstedt, R. H., 1997. Regulation of mem- brane stability and the acrosome reaction in mammalian sperm., 11 (8): 670-682.
Nonaka, S., Tanaka, Y., Okada, Y., Takeda, S., Harada, A., Kanai, Y.,., 1998. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein., 95 (6): 829-837.
Nuurai, P., Panasophonkul, S., Tinikul, Y., Sobhon, P., and Wani- chanon, R., 2016. Spermatogenesis in the rock oyster,(Gmelin, 1791)., 48 (1): 43-48.
O’Donnell, L., and O’Bryan, M. K., 2014. Microtubules and sper- matogenesis., 30: 45-54.
Pampliega, O., Orhon, I., Patel, B., Sridhar, S., Diaz-Carretero, A., Beau, I.,., 2013. Functional interaction between auto- phagy and ciliogenesis., 502 (7470): 194-200.
Pan, X., Ou, G., Civelekogluscholey, G., Blacque, O. E., Endres, N. F., Tao, L.,., 2006. Mechanism of transport of IFT par- ticles incilia by the concerted action of kinesin-II and OSM-3 motors., 174 (7): 1035-1045.
Prevo, B., Scholey, J. M., and Peterman, E. J. G., 2017. Intra- flagellar transport: Mechanisms of motor action, cooperation, and cargo delivery., 284 (18):2905-2931.
Ramalhosantos, J., Varum, S., Amaral, S., Mota, P. C., Sousa, A. P., and Amaral, A., 2009. Mitochondrial functionality in repro- duction: From gonads and gametes to embryos and embryonic stem cells., 15 (5): 553-572.
Rashid, D. J., Wedaman, K, P., and Scholey, J. M., 1995. Heterodi- merization of the two motor subunits of the heterotrimeric ki- nesin, KRP(85/95)., 252 (2): 157- 162.
Rattner, J. B., and Brinkley, B. R., 1972. Ultrastructure of mam- malian spermiogenesis. 3. The organization and morphogenesis of the manchette during rodent spermiogenesis., 41 (3):209-218.
Rosenbaum, J. L., and Witman, G. B., 2002. Intraflagellar trans- port., 3 (11): 813-825.
Rupik, W., Huszno, J., and Klag, J., 2011. Cellular organisation of the mature testes and stages of spermiogenesis in(Cyprinidae; Teleostei)–structural and ultrastructural studies., 42 (8):833.
Russell, L. D., Russell, J. A., MacGregor, G. R., and Meistrich, M. L., 1991. Linkage of manchette microtubules to the nuclear envelope and observations of therole of the manchette in nu- clear shaping during spermiogenesis in rodents., 192 (2): 97-120.
Saade, M., Irla, M., Govin, J., Victorero, G., Samson, M., and Ngu-yen, C., 2007. Dynamic distribution of spatial during mouse spermatogenesis and its interaction with the kinesin KIF17b., 313 (3): 614-626.
Schimert, K. I., Budaitis, B. G., Reinemann, D. N., Lang, M. J., and Verhey, K. J., 2019. Intracellular cargo transport by single- headed kinesin motors., 116 (13): 6152-6161.
Scholey, J. M., 2012. Kinesin-2 motors transport IFT-particles, dy-neins and tubulin subunits to the tips ofsensory cilia: Relevance to vision research?, 75: 44-52.
Shen, H. Q., Xiao, Y. X., She, Z. Y., Tan, F. Q., and Yang, W. X., 2017. A novel role of KIF3b in the seminoma cell cycle., 352 (1): 95-103.
Singson, A., 2001. Every sperm is sacred: Fertilization in., 230 (2): 101-109.
Stauber, T., Simpson, J. C., and Pepperkok, R., 2006. A role for kinesin-2 in COPI-dependent recycling between the ER and the golgi complex., 16 (22): 2245-2251.
Su, X. R., Du, L. L., Li, Y. Y., Li, Y., Zhou, J., and Li, T., 2010. Cloning and expression of HSP70 gene ofsipuncula, 28 (3): 461- 466.
Wang, J. Q., Gao, X. M., Zheng, X. B., Hou, C. C., Xie, Q. P., Lou, B.,., 2019. Expression and potential functions of KIF3A/ 3B to promote nuclear reshaping and tail formation duringspermiogenesis., 229 (5-6): 161-181.
Wang, W., Dang, R., Zhu, J. Q., and Yang, W. X., 2010. Identi- fication and dynamic transcription of KIF3A homologue gene in spermiogenesis of., 157 (3): 237-245.
Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N., 1994. KIF3B forms a heterodimer with KIF3A and works as a new microtubule-based anterograde motor of membrane organelle transport., 19: S84.
Yamazaki, H., Nakata, T., Okada, Y., and Hirokawa, N., 1996. Clo- ning and characterization of KAP3: A novel kinesin superfami- ly-associated protein of KIF3A/3B., 93 (16): 8443-8448.
Yang, W. X., and Sperry, A. O., 2003. C-terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle trans- port., 69 (5): 1719-1729.
Yang, Z., Roberts, E. A., and Goldstein, L. S., 2001. Functional analysis of mouse kinesin motor Kif3C., 21 (16): 5306-5311.
Yu, K., Hou, L., Zhu, J. Q., Ying, X. P., and Yang, W. X., 2009. KIFC1 participates in acrosomal biogenesis, with discussion ofits importance for the perforatorium in the Chinese mitten crab., 337 (1): 113-123.
Zhang, Z. G., Li, W., Zhang, Y., Zhang, L., Teves, M., Liu, H.,.,2016. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice.,27 (23):3705-3716.
Zhao, Y. Q., Mu, D. L., Wang, D., Han, Y. L., Hou, C. C., and Zhu, J. Q., 2018. Analysis of the function of KIF3A and KIF3B in the spermatogenesis in., 44 (3): 769-788.
Zhao, Y. Q., Yang, H. Y., Zhang, D. D., Han, Y. L., Hou, C. C., and Zhu, J. Q., 2017. Dynamic transcription and expression patterns of KIF3A and KIF3B genes during spermiogenesis in the shrimp,., 184: 59-77.
Zhu, J. Q., Wang, W., Xu, S. J., and Zeng, H. X., 2007. Sperma- togenesis and sperm morphology of., 53 (4): 733-741.
December 15, 2020;
March 22, 2021;
July 6, 2021
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
. E-mail: zhujunquan@nbu.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2022年4期
Journal of Ocean University of China2022年4期
- Journal of Ocean University of China的其它文章
- The Role of Bottom Currents on the Morphological Development Around a Drowned Carbonate Platform, NW South China Sea
- Controls on the Gas Hydrate Occurrence in Lower Slope to Basin-Floor, Northeastern Bay of Bengal
- Effective Elastic Thickness of the Lithosphere in the Mariana Subduction Zone and Surrounding Regions and Its Implications for Their Tectonics
- Geophysical Evidence for Carbonate Platform Periphery Gravity Flows in the Xisha Islands, South China Sea
- Seismic Characteristics and Hydrocarbon Accumulation Associated with Mud Diapir Structures in a Superimposed Basin in the Southern South China Sea Margin
- Simulation and Analysis of Back Siltation in a Navigation Channel Using MIKE 21
