原核细胞精氨酸生物合成途径的研究进展
闫洪波王威李令娣安万昌
(1.滨州生物技术研究院,滨州 256600;2.山东民强生物科技股份有限公司,滨州 256600)
原核细胞精氨酸生物合成途径的研究进展
闫洪波1,2王威2李令娣2安万昌2
(1.滨州生物技术研究院,滨州 256600;2.山东民强生物科技股份有限公司,滨州 256600)
原核生物的精氨酸生物合成包含8个酶系,起始于乙酰谷氨酸激酶催化的谷氨酸的乙酰化。到第五步乙酰基团脱离,乙酰谷氨酸通过3个酶的作用,进一步合成乙酰化中间产物。鸟氨酸被氨甲酰基化生成瓜氨酸,天冬氨酸介入后形成精氨琥珀酸,最后形成终产物精氨酸。主要就精氨酸生物合成途径、合成过程中主要酶系及反馈抑制蛋白的作用机制进行了概述。此外,提出了目前精氨酸代谢研究中存在的问题及未来的研究方向。
精氨酸;乙酰谷氨酸合成酶;乙酰谷氨酸激酶;鸟氨酸乙酰基转移酶;ArgR
DIO: 10.13560/j.cnki.biotech.bull.1985.2015.01.003
目前,生物发酵的氨基酸工业化生产主要是通过控制微生物的代谢合成过程来实现,涉及微生物代谢网络和调控机制。在精氨酸的微生物发酵过程中,它既是目标产物处于代谢链的下游,也是腐胺、多胺和精胺的合成前体,这些聚胺类物质涉及几个生理过程是机体最佳生长所必须的。此外,当氮源缺乏条件下,通过精氨酸琥珀酰基转移酶的作用精氨酸可充当微生物生长的氮源[1,2]。目前,国内外尚无关于L-精氨酸生物合成途径的专一综述性论文。因此,本文对精氨酸生物合成途径的研究概述以期为精氨酸发酵工艺控制与优化、发酵菌种的定向选育及基因工程菌株的构建提供理论指导;开拓L-精氨酸的应用研究的思路,以及为其在临床医疗和运动保健等方面的应用提供理论依据。
1 精氨酸的应用
高血氨症是一个临床难题,可严重影响中枢神经系统,主要由肝脏疾病或者遗传性新陈代谢紊乱引起。L-精氨酸是生物体内的一种半必需碱性氨基酸,是尿素循环途径中重要代谢产物之一,可把高浓度的氨转换为尿素随尿液排出,从而有效解除血氨中毒[3]。L-精氨酸是一氧化氮的自然前体物质,在一氧化氮信号途径中通过一氧化氮合酶(Nitricoxide synthase,NOS)的催化生成一氧化氮和瓜氨酸,从而具有松弛血管平滑肌的功能[4-7]。此外,其抗肿瘤、治疗肾衰竭等免疫功能也成为研究热点[8-10]。因此,L-精氨酸在医疗、保健等领域具有广泛应用。
2 精氨酸生物合成途径概述
在原核细胞内,精氨酸的生物合成是涉及连续的8个酶的催化过程,起始于L-谷氨酸的乙酰化作用,这个反应是通过乙酰谷氨酸合成酶(N-acetylglutamate synthase NAGS)催化完成,精氨酸合成的首要步骤要防止谷氨酸环化及进一步合成脯氨酸[11]。精氨酸起始合成涉及乙酰谷氨酸是大多数原核生物、真菌、植物(包括单细胞藻类)的重要特征[12]。在乙酰谷氨酸合成之后,通过3个酶催化进一步合成乙酰化中间产物,该过程直到第五步乙酰基团脱除生成鸟氨酸。鸟氨酸氨甲酰基化生成瓜氨酸,而天冬氨酸介入合成了精氨琥珀酸,继而生成终产物精氨酸[13]。鉴于在所有生物体中精氨酸代谢的重要性以及哺乳动物体内精氨酸合成并未涉及乙酰化的中间产物[14,15],微生物和植物的精氨酸合成途径提供了一种潜在的抗菌或灭菌复合物的开发机制[16]。
由乙酰基团脱离方式的差异,可分为2条不同的精氨酸生物合成途径。有些细菌(肠杆菌科Enterobacteriaceae、弧菌科Vibrionaceae、黄色黏球菌Myxococcus xanthus和古菌类的硫化叶菌Sulfolobus solfataricus等)使用线性途径,其特征是通过乙酰鸟氨酸脱乙酰基酶水解乙酰鸟氨酸生成醋酸和鸟氨酸,在这些生物体内,合成路径第一个酶乙酰谷氨酸合成酶是精氨酸反馈抑制的靶点[17-20](图1-A);其他的细菌(奈瑟氏菌Neisseria spp.和链霉菌Streptomyces spp.)包括棒杆菌(Corynebacterium)使用更经济的乙酰基循环途径[21],通过鸟氨酸乙酰转移酶催化乙酰基团从乙酰鸟氨酸到谷氨酸转移,从而使乙酰谷氨酸再次循环进入精氨酸合成过程,此过程中受到精氨酸反馈抑制的是乙酰谷氨酸激酶(图1-C)。近几年,人们在黄单胞菌(Xanthomonas campestris)中发现一条新颖的精氨酸合成线性途径[22],在乙酰鸟氨酸氨甲酰基磷酸酶作用下,鸟氨酸的合成前体乙酰鸟氨酸直接转换为乙酰瓜氨酸,并且推断乙酰鸟氨酸脱乙酰基酶作用于乙酰瓜氨酸,使之脱除乙酰基,生成瓜氨酸(图1-B)。
3 精氨酸生物合成的酶系与基因
在某些细菌(假单胞菌Pseudomonas spp.和蓝藻菌Cyanobacterium spp.)中,精氨酸生物合成基因是分散在染色体上的[23,24],而在另外一些细菌(结核分枝杆菌M. tuberculosis和谷氨酸棒杆菌C. glutamicum)中是成簇集中的[25,26]。谷氨酸棒杆菌中存在由8个结构基因组成的精氨酸生物合成基因簇argCJBDFRGH,其分别从argC和argG的启动子开始独立转录,从而产生与之对应的两个有序排列的操纵子argCJBDFR和argGH[26,27],可编码从谷氨酸到精氨酸代谢合成必需的所有酶系(图2-A)。
3.1 乙酰谷氨酸合成酶(NAGS)在精氨酸生物合成中的作用机制
乙酰谷氨酸(N-acetylglutamate,NAG)是哺乳动物细胞内肝脏尿素合成所必需的酶辅因子,同时也是微生物和植物细胞内精氨酸生物合成途径最初的中间产物,这一过程是在乙酰辅酶A存在条件下,由乙酰谷氨酸合成酶(NAGS)催化底物谷氨酸完成[28]。采用线性途径合成精氨酸的细菌或植物细胞内,NAGS受到精氨酸的反馈抑制,而哺乳动物细胞内NAGS受到精氨酸的正向调节[29,30]。
在大多数微生物中,精氨酸合成途径的第一步是通过NAGS催化的,人们首先在大肠杆菌中发现了NAGS的活性[31],它是一个同源六聚体,其中280个残基的N端被称为氨基酸激酶域(Amino acid kinase domain,AAK),类似于乙酰谷氨酸激酶(NAGK)[32-34],还具有150个残基的C端GNAT域[29,33],这两个结合域分别是作为精氨酸反馈抑制和催化底物的结合位点[33]。在NAGS中,3个AAK域二聚物通过N端的螺旋连接成同一的六聚环状,相邻AAK域二聚物均带有一个GNAT位点[33]。在绿脓杆菌(Pseudomonas aeruginosa)中,通过改变上述两个结构域间“短连接器”的长度与顺序可以显著降低甚至消除精氨酸对于NAGS的抑制作用,恢复适当酶活性,这个“连接器”在精氨酸的调控中具有至关重要的作用[34]。
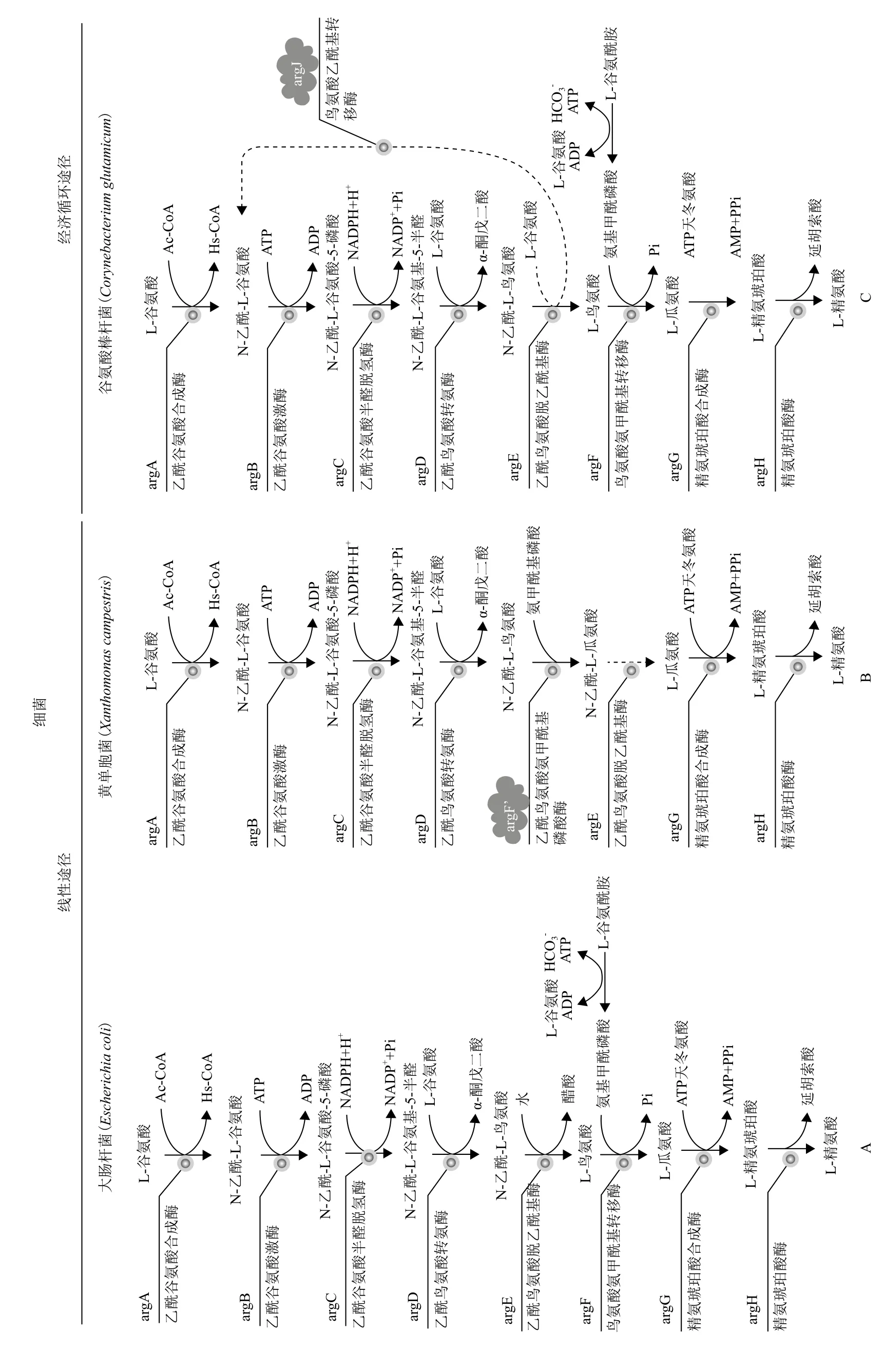
图1 原核细胞精氨酸生物合成途径[20-22]
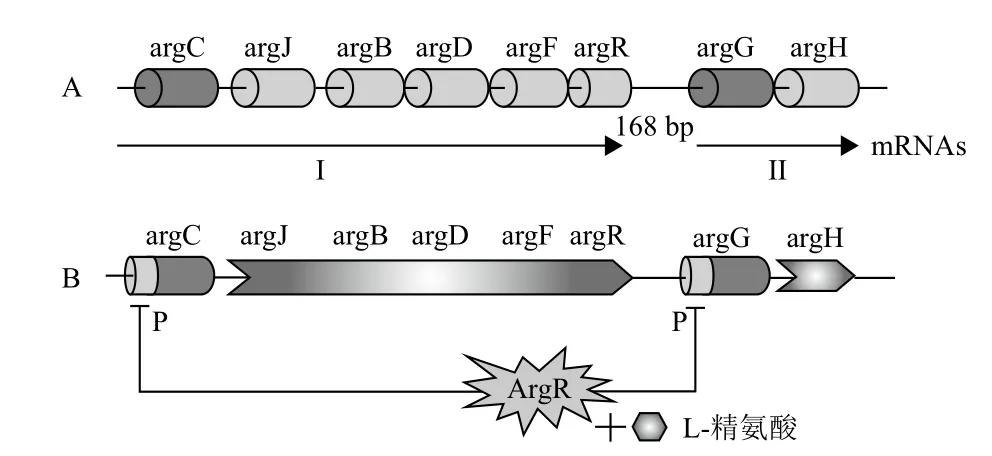
图2 精氨酸生物合成基因簇及ArgR蛋白对其调控机制[26,27]
3.2 乙酰谷氨酸激酶(NAGK)在精氨酸合成中的作用机制
乙酰谷氨酸激酶(N-acetyl-L-glutamate kinase,NAGK)是氨基酸激酶家族酶系之一,利用ATP催化乙酰谷氨酸(N-acetyl-L-glutamate,NAG)的γ-COO-群组磷酸化生成无水酰基磷酸盐,且这个过程具有可逆性[35,36]。大肠杆菌的NAGK具有氨基酸家族最典型的结构特征,27 kD亚基的同型二聚体高分辨率结构被测定,复合物带有亚胺二磷酸腺苷(AMPPNP)和NAG基质(PDB,1GS5)或者带有二磷酸腺苷(ADP)和硫酸根(PDB,1OHB)[16,35,36]。此外,嗜热细菌(Thermus thermophilus)、海栖热袍菌(Thermotoga maritima)和绿脓杆菌(Pseudomonas aeruginosa)的NAGK晶体结构也相继被测定[37,38],这对于深入了解NAGK在精氨酸合成途径中的作用奠定了重要的理论基础。
精氨酸生物合成的经济循环途径中,argB编码的乙酰谷氨酸激酶NAGK是精氨酸生物合成一个关键酶,其受到精氨酸的反馈抑制和反馈阻遏[39]。谷氨酸起始的下游代谢主要包括脯氨酸、谷氨酰胺和精氨酸3个途径。ChIP分析显示,添加代谢物脯氨酸,特异性减弱了ArgR与argB上游的结合能力,影响argB基因表达水平,导致鸟氨酸合成代谢增强,这表明argB上游区域在ArgR蛋白和arg操纵子的相互作用中起重要作用[40]。另有试验证明,鸟氨酸合成代谢受到ArgR调控,但其反馈抑制调控依赖于其与arg操纵子的DNA结合能力而不是ArgR蛋白的表达水平[41]。近期研究表明,谷氨酸棒杆菌ArgR蛋白在argB启动子区的结合位点是-77到-25,而另一个已被证明也可以结合到argB启动子区域的转录抑制子FarR在argB启动子区结合位点是-57到-77,二者结合域恰好重叠。一个值得注意的现象是,如果ArgR先结合到argB启动子区-77到-25位置,那FarR将无法结合到argB的启动子区域,相反,若FarR先结合到argB启动子区的-57到-77位置,那ArgR将转移结合到argB启动子区-49到-25位置,这表明在NAGK的反馈抑制过程中两个调控蛋白间也具有相互作用[42]。
3.3 鸟氨酸乙酰基转移酶(OAT)在精氨酸生物合成中的作用机制
鸟氨酸乙酰基转移酶(Ornithine acetyltransferase,OAT)是由argJ基因编码,最初在人类致病菌奈瑟菌(Neisseria gonorrhoeae)中被检测并进行了基因克隆与序列分析[43-45]。采用经济循环途径合成精氨酸的原核生物体内,OAT兼具谷氨酸合成酶NAGS与乙酰鸟氨酸脱乙酰基酶(N-acety-L-ornithine deacetylase,NAO)的功能,在乙酰基团的循环利用过程中发挥着重要作用,同时argJ编码的OAT酶会受到鸟氨酸的反馈抑制[39]。随后多个物种的OAT酶被研究,涉及编码基因及序列分析、酶学功能分析、动力学机制分析和系统进化理论等[12,39,46-48],有些物种的OAT酶不具有双重活性,它们无法利用作为乙酰基团来源的乙酰辅酶A[47]。
后续研究发现嗜热芽孢杆菌(Bacillus stearothermophilus)OAT具有双官能团,其活性来自于前体蛋白在高保守ATML序列的丙氨酸(A)和苏氨酸(T)残基间自我催化裂解,形成α亚基和β亚基后再聚合成一个异四聚体[48],β亚基已经证实在乙酰基转移反应中可形成一个乙酰基中间产物[48]。通过定点突变技术证实Thr97在OAT前体蛋白自我催化裂解而具有催化活性这一过程内具有重要作用[48]。在经济循环途径中,大多数的乙酰谷氨酸主要是由OAT催化谷氨酸和乙酰鸟氨酸产生,这种情况下,少量的NAGS在精氨酸合成过程中起到必要的补充功能[11]。
4 ArgR蛋白在L-精氨酸生物合成中起负调控作用
早期对于ArgR的研究主要围绕大肠杆菌(E. coli)、酿酒酵母(Saccharomyces cerevisiae)和鼠伤寒沙门氏菌(Salmonella typhimurium)开展[49-51],随后的研究证实ArgR蛋白及其调控机制在其他多个生物体中保守存在,如绿脓杆菌(Pseudomonas aeruginosa)、谷氨酸棒杆菌(C. glutamicum)、天蓝链霉菌(Streptomyces coelicolor)和结核分枝杆菌(Mycobacterium tuberculosis)等[41,52-54]。
ArgR蛋白是由argR基因编码,作为精氨酸生物合成的抑制子,其协同L-精氨酸负调控精氨酸代谢途径中酶系的合成[55]。在大肠杆菌(E.coli)中,ArgR是一个典型的反馈调节器,可有效传导胞内L-精氨酸信号,二者结合成为一个共抑制子[56,57]。ArgR不同于细菌里其他抑制子,它以相同16.5 kD多肽构成的六聚体形式发挥功能[55,57],大肠杆菌(E.coli)精氨酸生物合成的8个结构基因的启动子区域都具有两个相邻的18 bp的保守回文序列,这是ArgR结合的靶标位点,通常称为ARG盒[58,59]。
谷氨酸棒杆菌的ArgR蛋白也是一个同等亚基聚合而成的六聚体,蛋白分子量为110 kD。对ArgR蛋白序列分析显示,在N端存在一个“SR”(Ser57-Arg58)基序,这是一个保守的转录调控的DNA结合位点;同时在C端存在一个“GTIAGDDTV”基序,是L-精氨酸的结合位点,其中“DD”残基是已知的对ArgR蛋白有充分低聚作用[27]。通过体内染色质免疫沉淀(ChIP)分析,证实了ArgR蛋白与argC、argJ、argB、argD、argF、argR、argG和argH基因上游操纵子位点的相互作用[40]。通过argR基因过表达和基因敲除,证实了谷氨酸棒杆菌的ArgR抑制子在argC和argG的启动子上的作用(图2-B)。通过lacZ活性和argR突变分析,L-精氨酸存在的条件下,ArgR作用于argC的启动子来调控精氨酸生物合成基因簇argCJBDFR的转录。凝胶迁移(EMSA)试验显示ArgR的六聚体可直接绑定在argC的启动子上,也证实了上述结果,但是argGH的转录并未响应argR抑制和精氨酸的水平[27]。
5 展望
L-精氨酸涉及NO途径和尿素循环途径,同时也是多个蛋白和氨基酸的前体物质,因此被广泛用于医药和保健品的开发。精氨酸生物发酵是以微生物的代谢途径为基础,通过人工诱变和基因工程改造等手段减弱或消除终产物的反馈抑制,从而实现规模化生产。通过对多个生物体内精氨酸生物合成途径的研究,目前对于精氨酸的合成代谢途径了解较为清晰,这为基因工程菌株的构建提供了重要的理论依据[13,27,60]。我国用来产精氨酸的菌种多为谷氨酸棒杆菌、钝齿棒杆菌和黄色短杆菌的人工诱变菌株[61,62],因存在产酸量较低,发酵时间长、糖酸转化率较低和污染严重等问题,使规模化生产受困,而定向基因工程改造可以作为解决这些问题的一种有效手段。其次,对于L-精氨酸的对合成代谢过程的反馈调节机制的研究需要进一步深化,特别是L-精氨酸对NAGK的反馈抑制、L-鸟氨酸对OAT的反馈抑制、ArgR蛋白对于精氨酸生物合成基因簇的调控机制,以及该蛋白在生物体其他代谢途径中的功能与作用的研究对于有效解除L-精氨酸的反馈抑制具有重要的理论指导意义。
[1]Schneider BL, Kiupakis AK, Reitzer LJ. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli[J]. Journal of Bacteriology, 1998, 180(16):4278-4286.
[2]Lu CD. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains[J]. Applied Microbiology and Biotechnology, 2006, 70(3):261-272.
[3]Rajagopal BS, Deponte J, Tuchman M, et al. Use of inducible feedback-resistant N-acetylglutamate synthetase(argA)genes for enhanced arginine biosynthesis by genetically engineered Escherichia coli K-12 strains[J]. Applied and Environmental Microbiology, 1998, 64(5):1805-1811.
[4]Ikeda M, Mitsuhashi S, Tanaka K, et al. Reengineering of a Corynebacterium glutamicum L-arginine and L-citrulline producer[J]. Applied and Environmental Microbiology, 2009, 75(6):1635-1641.
[5]Appleton J. Arginine:clinical potential of a semi-essential amino acid[J]. Alternative Medicine Review, 2002, 7(6):512-522.
[6]Haynes JJr, Baliga BS, Obiako B, et al. Zileuton induces hemoglobin F synthesis in erythroid progenitors:role of the L-arginine-nitric oxide signaling pathway[J]. Blood, 2004, 103(10):3945-3950.
[7]Olinto SC, Adriao MG, Castro-Barbosa T, et al. Arginine induces GH gene expression by activating NOS/NO signaling in rat isolated hemi-pituitaries[J]. Brazilian Journal of Medical and Biological Research,2012, 45(11):1066-1073.
[8]Gonzaga Silva LF, Odorico de Moraes M, Santos Dias Soares F, et al. Effects of L-arginine-enriched total enteral nutrition on body weight gain, tumor growth, and in vivo concentrations of blood and tissue metabolites in rats inoculated with Walker tumor in the kidney[J]. Nutrition, 2004, 20(2):225-229.
[9]Suliburska J, Bogdanski P, Krejpcio Z, et al. The effects of L-arginine,alone and combined with vitamin C, on mineral status in relation to its antidiabetic, anti-inflammatory, and antioxidant properties in male rats on a high-fat diet[J]. Biological Trace Element Research,2014, 157(1):67-74.
[10]Nesher N, Frolkis I, Schwartz D, et al. L-Arginine improves endothelial function, independently of arginine uptake, in aortas from chronic renal failure female rats[J]. American Journal of Physiology Renal Physiology, 2014, 306(4):F449-456.
[11]Caldovic L, Tuchman M. N-acetylglutamate and its changing role through evolution[J]. The Biochemical Journal, 2003, 372(Pt 2):279-290.
[12]Xu Y, Labedan B, Glansdorff N. Surprising arginine biosynthesis:a reappraisal of the enzymology and evolution of the pathway in microorganisms[J]. Microbiology and Molecular Biology Reviews, 2007, 71(1):36-47.
[13] Petri K, Walter F, Persicke M, et al. A novel type of N-acetylglutamate synthase is involved in the first step of arginine biosynthesis in Corynebacterium glutamicum[J]. BMC Genomics, 2013, 14:713.
[14]Jones ME. Catalysts of the urea cyclea[J]. Transactions of the New York Academy of Sciences, 1983, 41(1 Series II):77-82.
[15]Alonso E, Rubio V. Participation of ornithine aminotransferase in the synthesis and catabolism of ornithine in mice. Studies using gabaculine and arginine deprivation[J]. The Biochemical Journal, 1989, 259(1):131-138.
[16]Gil-Ortiz F, Ramon-Maiques S, Fernandez-Murga ML, et al.Two crystal structures of Escherichia coli N-acetyl-L-glutamate kinase demonstrate the cycling between open and closed conformations[J]. Journal of Molecular Biology, 2010, 399(3):476-490.
[17] Cunin R, Glansdorff N, Piérard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria[J]. Microbiol Rev, 1986, 50(3):314-352.
[18]Harris BZ, Singer M. Identification and characterization of the Myxococcus xanthus argE gene[J]. Journal of Bacteriology,1998, 180(23):6412-6414.
[19]Van de Casteele M, Legrain C, Desmarez L, et al. Pathways of arginine biosynthesis in extreme thermophilic archaeo and eubacteria[J]. Journal of General Microbiology, 1990, 136:1177-1183.
[20]Xu Y, Liang ZY, Legrain C, et al. Evolution of arginine biosynthesis in the bacterial domain:novel gene-enzyme relationships from psychrophilic Moritella strains(Vibrionaceae)and evolutionary significance of N-alpha-acetyl ornithinase[J]. Journal of Bacteriology, 2000, 182(6):1609-1615.
[21] Udaka S. Pathway-specific pattern of control of arginine biosynthesis in bacteria[J]. Journal of Bacteriology, 1966, 91(2):617-621.
[22] Qu Q, Morizono H, Shi D, et al. A novel bifunctional N-acetylglutamate synthase-kinase from Xanthomonas campestris that is closely related to mammalian N-acetylglutamate synthase[J]. BMC Biochemistry, 2007, 8:4.
[23] Haas D, Holloway BW, Schambock A, et al. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa[J]. Molecular & General Genetics, 1977, 154(1):7-22.
[24]Floriano B, Herrero A, Flores E. Analysis of expression of the argC and argD genes in the cyanobacterium Anabaena sp. strain PCC 7120[J]. Journal of Bacteriology, 1994, 176(20):6397-6401.
[25] Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence[J]. Nature, 1998, 393(6685):537-544.
[26]Jiao H. Analysis of the arginine biosynthetic gene cluster argCJBDFR of Corynebacterium crenatum[J]. Journal of Biomedical Science and Engineering, 2011, 4(1):70-75.
[27] Yim SH, Jung S, Lee SK, et al. Purification and characterization of an arginine regulatory protein, ArgR, in Corynebacterium glutamicum[J]. Journal of Industrial Microbiology & Biotechnology, 2011,38(12):1911-1920.
[28]Caldovic L, Ah Mew N, Shi D, et al. N-acetylglutamate synthase:structure, function and defects[J]. Molecular Genetics and Metabolism, 2010, 100(Suppl 1):S13-19.
[29]Sancho-Vaello E, Fernandez-Murga ML, Rubio V. Site-directedmutagenesis studies of acetylglutamate synthase delineate the site for the arginine inhibitor[J]. FEBS Letters, 2008, 582(7):1081-1086.
[30]Cunin R, Glansdorff N, Pierard A, et al. Biosynthesis and metabolism of arginine in bacteria[J]. Microbiol Rev, 1986, 50(3):314-352.
[31]Maas WK, Novelli GD, Lipmann F. Acetylation of glutamic acid by extracts of Escherichia coli[J]. Proceedings of the National Academy of Sciences of the United States of America, 1953, 39(10):1004-1008.
[32]Parra-Gessert L, Koo K, Fajardo J, et al. Processing and function of a polyprotein precursor of two mitochondrial proteins in neurospora crassa[J]. The Journal of Biological Chemistry, 1998, 273(14):7972-7980.
[33]Sancho-Vaello E, Fernandez-Murga ML, Rubio V. Mechanism of arginine regulation of acetylglutamate synthase, the first enzyme of arginine synthesis[J]. FEBS Letters, 2009, 583(1):202-206.
[34]Sancho-Vaello E, Fernandez-Murga ML, Rubio V. Functional dissection of N-acetylglutamate synthase(ArgA)of Pseudomonas aeruginosa and restoration of its ancestral N-acetylglutamate kinase activity[J]. Journal of Bacteriology, 2012, 194(11):2791-2801.
[35]Gil-Ortiz F, Ramón-Maiques S, Fita I, et al. The course of phosphorus in the reaction of N-acetyl-l-glutamate kinase,determined from the structures of crystalline complexes, including a complex with an AlF(4)(-)transition state mimic[J]. Journal of Molecular Biology, 2003, 331(1):231-244.
[36]Marco-Marín C, Ramón-Maiques S, Tavárez S, et al. Sitedirected mutagenesis of Escherichia coli acetylglutamate kinase and aspartokinase III probes the catalytic and substrate-binding mechanisms of these amino acid kinase family enzymes and allows three-dimensional modelling of aspartokinase[J]. Journal of Molecular Biology, 2003, 334(3):459-476.
[37] Ramon-Maiques S, Fernandez-Murga ML, Gil-Ortiz F, et al. Structural bases of feed-back control of arginine biosynthesis, revealed by the structures of two hexameric N-acetylglutamate kinases, from Thermotoga maritima and Pseudomonas aeruginosa[J]. Journal of Molecular Biology, 2006, 356(3):695-713.
[38] Sundaresan R, Ragunathan P, Kuramitsu S, et al. The structure of putative N-acetyl glutamate kinase from Thermus thermophilus reveals an intermediate active site conformation of the enzyme[J]. Biochemical and Biophysical Research Communications, 2012, 420(3):692-697.
[39]Baetens M, Legrain C, Boyen A, et al. Genes and enzymes of the acetyl cycle of arginine biosynthesis in the extreme thermophilic bacterium Thermus thermophilus HB27[J]. Microbiology, 1998,144(Pt 2):479-492.
[40]Lee SY, Shin HS, Park JS, et al. Proline reduces the binding of transcriptional regulator ArgR to upstream of argB in Corynebacterium glutamicum[J]. Applied Microbiology and Biotechnology, 2010,86(1):235-242.
[41]Lee SY, Kim YH, Min J. The effect of ArgR-DNA binding affinity on ornithine production in Corynebacterium glutamicum[J]. Current Microbiology, 2009, 59(4):483-488.
[42]Lee SY, Park JM, Lee JH, et al. Interaction of transcriptional repressor ArgR with transcriptional regulator FarR at the argB promoter region in Corynebacterium glutamicum[J]. Applied and Environmental Microbiology, 2011, 77(3):711-718.
[43]Martin PR, Mulks MH. Molecular characterization of the argJ mutation in Neisseria gonorrhoeae strains with requirements for arginine, hypoxanthine, and uracil[J]. Infection and Immunity,1992, 60(3):970-975.
[44]Martin PR, Mulks MH. Sequence analysis and complementation studies of the argJ gene encoding ornithine acetyltransferase from Neisseria gonorrhoeae[J]. Journal of Bacteriology, 1992, 174(8):2694-2701.
[45]Shinners EN, Catlin BW. Arginine biosynthesis in Neisseria gonorrhoeae:enzymes catalyzing the formation of ornithine and citrulline[J]. Journal of Bacteriology, 1978, 136(1):131-135.
[46]De Rijcke M, Seneca S, Punyammalee B, et al. Characterization of the DNA target site for the yeast ARGR regulatory complex, a sequence able to mediate repression or induction by arginine[J]. Molecular and Cellular Biology, 1992, 12(1):68-81.
[47]Marc F, Weigel P, Legrain C, et al. Characterization and kinetic mechanism of mono- and bifunctional ornithine acetyltransferases from thermophilic microorganisms[J]. Eur J Biochem, 2000, 267(16):5217-5226.
[48] Marc F, Weigel P, Legrain C, et al. An invariant threonine is involved in self-catalyzed cleavage of the precursor protein for ornithine acetyltransferase[J]. The Journal of Biological Chemistry, 2001,276(27):25404-25410.
[49]Hirvonen AP, Vogel HJ. Response of argR- spheroplasts of Escherichia coli to extracted arginine repressor[J]. Biochemical and Biophysical Research Communications, 1970, 41(6):1611-1616.
[50]Hoet PP, Wiame JM. On the nature of argR mutations is Saccharomyces cerevisiae[J]. European Journal of Biochemistry / FEBS, 1974, 43(1):87-92.
[51]Kelln RA, Foltermann KF, O’Donovan GA. Location of the argR gene on the chromosome of Salmonella typhimurium[J]. Molecular & General Genetics, 1975, 139(4):277-284.
[52]Park SM, Lu CD, Abdelal AT. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons[J]. Journal of Bacteriology, 1997, 179(17):5309-5317.
[53]Cherney LT, Cherney MM, Garen CR, et al. Crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression[J]. Journal of Molecular Biology, 2010, 399(2):240-254.
[54]Perez-Redondo R, Rodriguez-Garcia A, Botas A, et al. ArgR of Streptomyces coelicolor is a versatile regulator[J]. PloS One,2012, 7(3):e32697.
[55]Lim DB, Oppenheim JD, Eckhardt T, et al. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product,the arginine repressor[J]. Proceedings of the National Academy of Sciences of the United States of America, 1987, 84(19):6697-6701.
[56]Maas WK. The arginine repressor of Escherichia coli[J]. Microbiol Rev, 1994, 58(4):631-640.
[57]Grandori R, Lavoie TA, Pflumm M, et al. The DNA-binding domain of the hexameric arginine repressor[J]. Journal of Molecular Biology, 1995, 254(2):150-162.
[58]Tian G, Lim D, Carey J, et al. Binding of the arginine repressor of Escherichia coli K12 to its operator sites[J]. Journal of Molecular Biology, 1992, 226(2):387-397.
[59]Charlier D, Roovers M, Van Vliet F, et al. Arginine regulon of Escherichia coli K-12. a study of repressor-operator interactions and of in vitro binding affinities versus in vivo repression[J]. Journal of Molecular Biology, 1992, 226(2):367-386.
[60] Xu M, Rao Z, Yang J, et al. Heterologous and homologous expression of the arginine biosynthetic argC~H cluster from Corynebacterium crenatum for improvement of(L)-arginine production[J]. Journal of Industrial Microbiology & Biotechnology, 2012, 39(3):495-502.
[61] Xu M, Rao Z, Dou W, et al. Site-directed mutagenesis and feedbackresistant N-acetyl-L-glutamate kinase(NAGK)increase Corynebacterium crenatum L-arginine production[J]. Amino Acids,2012, 43(1):255-266.
[62] Lv Y, Liao J, Wu Z, et al. Genome sequence of Corynebacterium glutamicum ATCC 14067, which provides insight into amino acid biosynthesis in coryneform bacteria[J]. Journal of Bacteriology,2012, 194(3):742-743.
(责任编辑 狄艳红)
Research Progress of the Arginine Biosynthetic Pathway in Prokaryotic Cells
Yan Hongbo1Wang Wei1,2Li Lingdi1,2An Wanchang2
(1.Binzhou Institute of Biotechnology,Binzhou 256600;2.Shandong Mingqing Biotechnology co.,LTD,Binzhou 256600)
Arginine biosynthesis in prokaryotes consists of eight enzymatic steps, starting with acetylation of glutamate, catalysed by N-acetylglutamate synthase(NAGS). After metabolisation of N-acetylglutamate, biosynthesis proceeds via three enzymatic steps which form further acetylated intermediates, until the acetyl group is removed in the fifth step of this process. The resulting ornithine is carbamoylated to citrulline. Addition of aspartate leads to N-argininosuccinate, which is finally converted to L-arginine. Here we summarize the arginine biosynthetic pathway, catalytic mechanisms of enzymes and the molecular mechanisms of feedback inhibition of ArgR protein. In addition, we briefly outline existing problems in the research of arginine metabolism and directions for future research.
arginine;NAGS;NAGK;OAT;ArgR
2014-04-18
山东省科技发展项目(2013GSF12118)
闫洪波,男,博士,研究方向:工业微生物基因工程改造;E-mail:hongbo_1981@126.com

