橡胶树树皮miRNA定量表达分析的内参筛选
吴绍华,张世鑫,杨署光,田维敏
橡胶树树皮miRNA定量表达分析的内参筛选
吴绍华,张世鑫,杨署光,田维敏*
中国热带农业科学院橡胶研究所/农业农村部橡胶树生物学与遗传资源利用重点实验室/省部共建国家重点实验室培育基地-海南省热带作物栽培生理学重点实验室,海南海口 571101
世界所需天然橡胶主要来自橡胶树,橡胶树的乳管是合成和储存天然橡胶的组织,树干树皮中的次生乳管与天然橡胶生产密切相关,由维管形成层分化而来。次生乳管数量与天然橡胶产量呈显著正相关。因此,乳管分化是天然橡胶生产面临的一个重大理论课题。植物miRNAs是一类长约20~24个核苷酸的非编码小RNA分子,通过介导基因沉默在植物生长发育、细胞分化及逆境适应中起着非常重要的调节作用。橡胶树乳管分化过程中差异miRNAs的鉴定对于进一步认识乳管分化的分子机理具有重要的作用。实时荧光定量PCR(qPCR)已广泛用于miRNA的定量表达分析,选择合适的miRNA内参对于准确进行miRNA的表达定量至关重要。本研究以冠菌素(coronatine, COR)诱导橡胶树萌条分化次生乳管的实验系统,采用小RNA Poly A加尾的qPCR技术分析COR处理对形成层区3个非编码RNA(、、)和6个miRNA(、、、、、)候选内参的表达稳定性的影响。geNorm和NormFinder软件的联合分析结果显示,表达稳定性最高的是和,稳定性较差的是。和可作为合适的内参基因,用于分析COR影响下的miRNA相对定量表达,为鉴定橡胶树次生乳管分化相关的差异表达miRNAs奠定良好基础。
橡胶树;乳管分化;miRNA;冠菌素;内参基因
天然橡胶作为四大工业原材料之一,在世界工业化及经济发展中起着重要的作用,是我国重要的战略物资。世界上98%的天然橡胶都来源于橡胶树(Muell. Arg.)。橡胶树的乳管是合成和贮存天然橡胶的组织,其数量与天然橡胶产量呈显著正相关。因此,乳管细胞的分化机理研究对于改良橡胶树的产胶潜力具有重要的意义。橡胶树树皮中的次生乳管是由维管形成层的纺锤状原始细胞分化而来。在前期的研究中,我们发现机械伤害、茉莉酸、冠菌素(coronatine, COR)及曲古抑菌素A(trichostatin A,TSA)均能诱导次生乳管的分化[1-5]。基于COR诱导次生乳管分化的实验系统,通过消减SSH文库及转录组,初步推测茉莉酸信号途径、CLAVATA-MAPK-WOX及钙调信号途径可能在次生乳管分化过程中起着重要的调控作用[4-6]。为了对这些信号途径的差异基因进行实行荧光定量PCR(qPCR)验证,通过COR和TSA诱导橡胶树萌条树皮乳管分化系统对22个候选的内参基因进行评估,结果显示在COR诱导次生乳管分化的实验系统中表达最稳定[6],而是TSA诱导橡胶树萌条次生乳管分化的过程中的最佳内参基因[7]。
miRNAs是一类长约21~24个核苷酸的非编码小RNA分子,其介导的基因表达调控在生物生长发育、细胞分化及适应各种逆境胁迫中起着非常重要的作用。目前,橡胶树miRNAs的研究主要集中在逆境响应[8-10]、胶乳代谢[11-13]、死皮相关miRNAs的鉴定[14-15]。对于miRNA介导橡胶树器官分化特别是次生乳管分化的研究未见报道。为研究miRNA介导次生乳管分化的调控机制,本课题组前期进行了COR诱导橡胶树内层树皮的小RNA高通量测序,初步获得了差异表达的miRNAs。为对差异miRNAs进行qPCR验证,需要筛选出合适的miRNA内参,用于miRNA定量表达分析。但有关COR诱导次生乳管分化过程中miRNA实时荧光定量PCR内参miRNA筛选未见报道。因此,本研究基于COR处理橡胶树内层树皮的小RNA高通量测序数据(未发表),筛选COR处理与对照表达量相对稳定的6个mature miRNA (、、、、、)和3个常用于其他作物miRNA定量的内参、和,采用geNorm[16]和NormFinder[17]软件进行评估,以筛选出适合橡胶树树皮COR响应过程中miRNA定量表达分析的内参。
1 材料与方法
1.1 材料
以橡胶树无性系‘热研7-33-97’萌条为实验材料。在自然条件下,每年新萌发的第1~2伸长单位的树皮是没有次生乳管的[2-3],COR诱导处理可产生次生乳管。本实验以第二伸长单位的树皮作为COR处理的部位。
1.2 方法
1.2.1 材料处理及含小RNA的总RNA提取 用单面刀片轻轻刮去第二伸长单位的茎表皮的角质层,然后分别涂20 μmol/L的冠菌素(Sigma, USA)包裹后处理1、2、8、24 h[4],于处理后采集树皮,每个时间点收集9株萌条的样品,液氮冷冻后于‒80℃冰箱保存,用于冰冻切片采集形成层区样品。冰冻切割的形成层区样品采用mirVana™ PARIS™ Kit (Invitrogen™, USA)试剂盒分离含小RNA的总RNA。采用DNA-free™ DNA去除试剂盒(Invitrogen™, USA)消化总RNA中的痕量DNA。RNA提取后采用琼脂糖凝胶电泳检测RNA的完整性,NanoDrop 2000 (Thermo Fisher Scientific,USA)测定RNA的纯度和浓度。
1.2.2 qPCR候选内参miRNA的选择 基于COR处理橡胶树内皮小RNA测序数据(未发表),筛选COR处理前后表达量相对稳定的6个miRNAs以及、、作为候选内参,候选内参的引物如表1。
1.2.3 小RNA的反转录及qPCR分析 本实验取1 µg RNA采用小RNA Poly(A)加尾反转录法,参照miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen,北京)说明书反转录成第一链cDNA。获得的cDNA产物稀释10倍后,参照miRcute增强型miRNA荧光定量检测试剂盒(SYBR Green, Tiangen,北京)说明书,基于CFX384 Real-Time PCR Detection System(Bio-Rad公司,USA)平台进行qPCR。

表1 本研究所用引物
1.3 数据处理
根据荧光定量结果,获得内参各样品的平均q值,按照公式将内参基因的原始q值转化为相对表达量值。
=Eq min‒Cq sample
式中,为基因的扩增效率,当扩增效率接近100%时,通常默认为2;q sample为该基因在各个组织中的q值,q min为该基因在所有组织中最小的q值。
然后将值导入geNorm和NormFinder软件中,对橡胶树COR响应内皮miRNA荧光定量PCR的内参进行稳定性的评估。geNorm分析根据值评估内参的稳定性,值<1.5。值与内参的稳定性呈负相关,即值越小基因越稳定。软件会根据值评估出最稳定的基因组合,当配对变异数V/n+1<0.15,只需要个内参进行定量;当V/n+1>0.15,则需要+1个内参对数据进行定量。
NormFinder软件通过计算基因的表达稳定值(stability value,)来评估候选内参基因的稳定性,值越小,候选内参就越稳定。最后,综合2个软件的评估结果筛选出适合橡胶树COR响应树皮miRNA荧光定量PCR的内参。
2 结果与分析
2.1 候选miRNA内参定量引物的特异性检测
采用RT-PCR对3个非编码RNA (、、)和6个miRNA (、、、、、)进行扩增,PCR产物经3%的琼脂糖凝胶电泳检测,结果显示,9个候选的miRNA内参扩增条带单一(图1),且荧光定量PCR产物的熔解曲线只有单一峰(图2),表明候选miRNA的内参的PCR产物单一、特异性好,符合qPCR实验标准,可用于内参miRNA基因的评估。
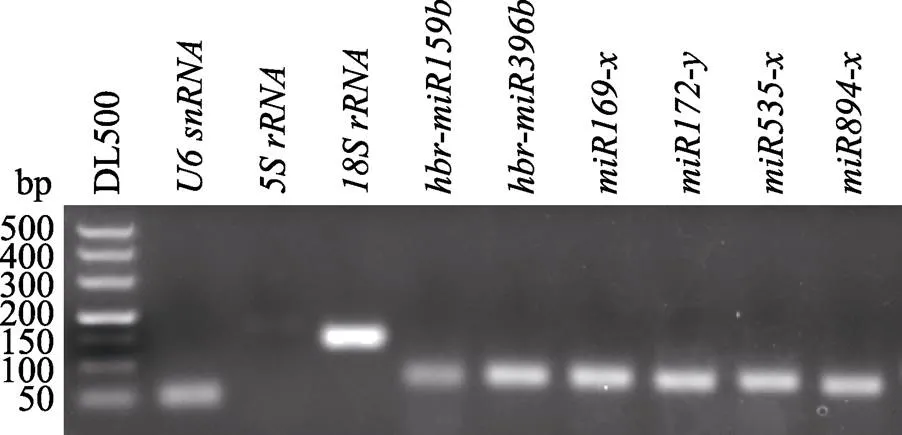
图1 橡胶树候选miRNA内参的RT-PCR扩增
2.2 COR处理和对照橡胶树萌条树皮形成层组织候选miRNA内参的转录丰度
本研究采用qPCR分析了3个非编码RNA和6个miRNA的转录丰度。根据荧光定量PCR熔解曲线分析显示,9个非编码RNA的PCR产物的熔解曲线均为单峰(图2),表明产物具有特异性,获得q值能准确有效地反映表达的丰度。q值预测内参基因的表达丰度,q值越小,表达丰度越高。根据候选miRNA内参表达的q值的分布情况,9个候选的miRNA内参的q值介于16.79~31.59之间,其中的q值最低,表达量最高;的q值最高,表达量最小。和6个miRNA的q值介于16.80~27.69之间(图3)。

图2 9个候选miRNA内参的qPCR熔解曲线

图3 9个候选miRNA内参的Cq值
2.3 geNorm分析候选miRNA内参表达的稳定性
geNorm分析结果显示,COR处理和对照橡胶树萌条形成层区组织候选miRNAs内参表达的稳定性从高到低依次为/>>>>>>>,表明橡胶树树皮对COR响应的过程中,前3组最稳定的miRNA分别为、、,其中和是最稳定的miRNA内参。最不稳定的小分子RNA是(图4A)。而候选miRNAs内参的配对差异值的分析结果显示,橡胶树树皮对COR响应的过程中,V3/4的配对的变异值(pairwise variations)最小(0.101),小于阈值0.15,可判定作为miRNA定量表达的最优内参个数为3个(图4B)。

A:geNorm软件评估的miRNA的表达稳定性平均值;B:候选miRNA内参的配对变异值。
2.4 NormFinder分析候选miRNA内参表达的稳定性
NormFinder分析结果显示,COR处理和对照橡胶树萌条形成层区组织中候选miRNAs内参表达的稳定性从高到低依次为>>>>>>>>,前3个最稳定的miRNA分别为、和,其中是最稳定的miRNA内参,最不稳定的小分子RNA是(图5)。综合NormFinder和geNorm分析结果显示,在稳定性前3位的miRNA中均包含和。而最不稳定的3位内参基因是一致的,表明橡胶树树皮对COR响应的过程中,和较适合作为miRNA相对定量表达的内参miRNA。
3 讨论
成熟miRNA的定量表达分析是初步进行miRNA功能鉴定的前提。miRNA的表达丰度的检测最初是采用Northern杂交和微阵列分析方法进行的。而stem loop qRT-PCR[18]和poly (A)-tailed qRT-PCR 荧光定量表达技术出现,使得miRNA这类较小的片段的检测变得灵敏、方便、快捷。但作为荧光定量表达技术,其准确性同样依赖于合适、稳定的内参基因的选择。目前,作为最常用的内参,用于诸如葡萄[19]、小桐子[20]、火龙果[21]、青花菜[22]等多种作物的miRNA定量表达的内参。但是,已有的研究证明,并没有绝对稳定的基因[23],在很多情况下的表达也是不稳定的,不适合作为内参,比如小麦[24]、龙眼[25]、核桃[26]等作物。在本研究中,我们通过geNorm与NomFinder软件评估,发现橡胶树树皮中的在COR处理条件下也是不稳定的,并不适合作为COR处理条件下橡胶树萌条树皮形成层区组织中miRNA的定量表达的内参。除了,、、和其他的miRNA也经常作为内参,但不同部位及不同处理条件,miRNA的内参都是不一致的。干旱胁迫下大豆根和叶中成熟miRNA定量的最适内参分别为和a[27]。miRNA表达的稳定性也受到了生物逆境胁迫的影响。在溃疡病菌感染下,是白杨木的最适内参[28];黄杆菌属subsp.侵染导致的柑桔溃疡病过程中,和是定量miRNA表达的最适内参[28]。在植物的发育过程中,miRNA定量的内参也是不一致的。甘蓝型油菜在种子的发育过程中miRNA表达分析的最适内参组合是、和[29];核桃不同分化期叶芽中miRNA表达分析的最佳内参为和[30];在青花菜花蕾发育过程中,和分别是花蕾4个不同部位以及花蕾不同发育时期的适宜内参基因[22]。因此,作为内参基因也是相对稳定的,为了较准确的定量miRNA的表达,应该评估出当前实验条件下的稳定表达的miRNA分子,以筛选出合适的miRNA内参。本研究采用geNorm与NomFinder软件评估COR诱导橡胶树次生乳管分化过程中稳定的miRNA,结果显示和较适合作为miRNA相对定量表达的内参miRNA,该研究结果将为进一步鉴定橡胶树次生乳管分化的差异表达miRNAs提供合适的内参选择。

图5 NormFinder软件评估COR处理橡胶树萌条形成层组织中9个miRNA候选内参表达的稳定性
[1] 郝秉中, 吴继林. 创伤(割胶)对巴西橡胶树乳管分化的影响[J]. 植物学报, 1982, 24(4): 388-392.
HAO B Z, WU J L. Effects of wound (tapping) on laticifer differentiation in[J]. Acta Botanica Sinica, 1982, 24(4): 388-392. (in Chinese)
[2] HAO B Z, WU J L. Laticifer differentiation in: induction by exogenous jasmonic acid and linolenic acid[J]. Annals of Botany, 2000, 85: 37-43.
[3] TIAN W M, YANG S G, SHI M J, ZHANG S X, WU J L. Mechanical wounding-induced laticifer differentiation in rubber tree: an indicative role of dehydration, hydrogen peroxide, and jasmonates[J]. Journal of Plant Physiology, 2015, 182: 95-103.
[4] ZHANG S X, WU S H, CHEN Y Y, TIAN W M. Analysis of differentially expressed genes associated with coronatine-induced laticifer differentiation in the rubber tree by subtractive hybridization suppression[J]. PLoS One, 2015, 10(7): e0132070.
[5] ZHANG S X, WU S H, TIAN W M. The secondary laticifer differentiation in rubber tree is induced by trichostatin A, an inhibitor of histone acetylation[J]. Frontiers of Agricultural Science and Engineering, 2016, 3(4): 357-362.
[6] WU S H, ZHANG S X, CHAO J Q, DENG X M, CHEN Y Y, SHI M J, TIAN W M. Transcriptome analysis of the signalling networks in coronatine-induced secondary laticifer differentiation from vascular cambia in rubber trees[J]. Scientific Reports, 2016, 6: 36384.
[7] 吴绍华, 张世鑫, 杨署光, 田维敏. TSA诱导橡胶树次生乳管分化过程qRT-PCR内参基因的筛选[J]. 热带作物学报, 2020, 41(3): 504-512.
WU S H, ZHANG S X, YANG S G, TIAN W M. Selection of reference genes for normalization of quantitative real-time PCR analysis in secondary laticifer differentiation induced by trichostatin a in rubber tree (Muell. Arg.)[J]. Chinese Journal of Tropical Crops, 2020, 41(3): 504-512. (in Chinese)
[8] GÉBELIN V, ARGOUT X, ENGCHUAN W, PITOLLAT B, DUAN C, MONTORO P, LECLERCQ J. Identification of novel microRNAs inand computational prediction of their targets[J]. BMC Plant Biology, 2012, 12: 18.
[9] GÉBELIN V, LECLERCQ J, HU S, TANG C, MONTORO P. Regulation of MIR genes in response to abiotic stress in[J]. International Journal of Molecular Sciences, 2013, 14(10): 19587-19604.
[10] ZHANG Y, LECLERCQ J, WU S, ORTEGA-ABBOUD E, POINTET S, TANG C, HU S, MONTORO P. Genome-wide analysis inlaticifers revealed species-specific post-transcriptional regulations of several redox-related genes[J]. Scientific Reports, 2019, 9(1): 5701.
[11] KANJANAWATTANAWONG S, TANGPHATSORNRUANG S, TRIWITAYAKORN K, RUANG-AREERATE P, SANGSRAKRU D, POOPEAR S, SOMYONG S, NARANGAJAVANA J. Characterization of rubber tree microRNA in phytohormone response using large genomic DNA libraries, promoter sequence and gene expression analysis[J]. Molecular Genetics and Genomics, 2014, 289(5): 921-933.
[12] PRAMOOLKIT P, LERTPANYASAMPATHA M, VIBOONJUN U, KONGSAWADWORAKUL P, CHRESTIN H, NARANGAJAVANA J. Involvement of ethylene-responsive microRNAs and their targets in increased latex yield in the rubber tree in response to ethylene treatment[J]. Plant Physiology and Biochemistry, 2014, 84: 203-212.
[13] LECLERCQ J, WU S, FARINAS B, POINTET S, FAVREAU B, VIGNES H, KUSWANHADI K, ORTEGA-ABBOUD E, DUFAYARD JF, GAO S, DROC G, HU S, TANG C, MONTORO P. Post-transcriptional regulation of several biological processes involved in latex production in[J]. Peer Journal, 2020, 8: e8932.
[14] GÉBELIN V, LECLERCQ J, KUSWANHADI, ARGOUT X, CHAIDAMSARI T, HU S, TANG C, SARAH G, YANG M, MONTORO P. The small RNA profile in latex fromtrees is affected by tapping panel dryness[J]. Tree Physiology, 2013, 33(10): 1084-1098.
[15] LIU H, HU Y, YUAN K, FENG C, HE Q, SUN L, WANG Z. Genome-wide identification of lncRNAs, miRNAs, mRNAs, and their regulatory networks involved in tapping panel dryness in rubber tree ()[J]. Tree Physiology, 2022, 42(3): 629-645.
[16] VANDESOMPELE J, DE PRETER K, PATTYN F, POPPE B, VAN ROY N, DE PAEPE A, SPELEMAN F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biology, 2002, 3(7): RESEARCH0034.
[17] ANDERSEN C L, JENSEN J L, ORNTOFT T F. Normalization of real-time quantitative reverse transcription-PCR data: amodel-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Research, 2004, 64(15): 5245- 5250.
[18] CHEN C, RIDZON D A, BROOMER A J, ZHOU Z, LEE D H, NGUYEN J T, BARBISIN M, XU N L, MAHUVAKAR V R, ANDERSEN M R, LAO K Q, LIVAK K J, GUEGLER K J. Real-time quantification of microRNAs by stem-loop RT-PCR[J]. Nucleic Acids Research, 2005, 33(20): e179.
[19] LUO M, GAO Z, LI H, LI Q, ZHANG C, XU W, SONG S, MA C, WANG S. Selection of reference genes for miRNA qRT-PCR under abiotic stress in grapevine[J]. Scientific Reports, 2018, 8(1): 4444.
[20] 孔春艳, 陈永坤, 王莎莎, 郝大海, 杨 宇, 龚 明. 小桐子低温胁迫下microRNA实时荧光定量PCR内参的筛选与比较[J]. 生物技术通报, 2019, 35(7): 25-31.
KONG C Y, CHEN Y K, WANG S S, HAO D H, YANG Y, GONG M. Screening and comparison of reference genes for microRNA quantitative real-time pcr inunder chilling stress[J]. Biotechnology Bulletin, 2019, 35(7): 25-31. (in Chinese)
[21] 李阿利, 杨 鹍, 文 壮, 仇志浪, 文晓鹏. 火龙果miRNA表达分析的qRT-PCR检测体系建立[J]. 种子, 2019, 38(3): 6-10, 14.
LI A L, YANG K, WEN Z, QIU Z L, WEN X P. Establishment of qRT-PCR detection system for miRNA expression in[J]. Seed, 2019, 38(3): 6-10, 14. (in Chinese)
[22] 裴徐梨, 荆赞革, 唐 征, 罗天宽. 青花菜花蕾发育miRNA荧光定量内参基因的筛选[J]. 基因组学与应用生物学, 2021, 40(Z1): 2201-2207.
PEI X L, JING Z G, TANG Z, LUO T K. Selection of miRNA reference genes for quantitative RT-PCR in developmental bud of broccoli[J]. Genomics and Applied Biology, 2021, 40(Z1): 2201-2207. (in Chinese)
[23] KULCHESKI F R, MARCELINO-GUIMARAES F C, NEPOMUCENO A L, ABDELNOOR R V, MARGIS R. The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean[J]. Analytical Biochemistry, 2010, 406(2): 185-192.
[24] FENG H, HUANG X L, ZHANG Q, WEI G R, WANG X J, KANG Z S. Selection of suitable inner reference genes for relative quantification expression of microRNA in wheat[J]. Plant Physiology and Biochemistry, 2012, 51: 116-122.
[25] LIN Y L, LAI Z X. Evaluation of suitable reference genes for normalization of microRNA expression by realtime reverse transcription PCR analysis during longan somatic embryogenesis[J]. Plant Physiology and Biochemistry, 2013, 66: 20-25.
[26] 周 丽, 全绍文, 马 丽, 徐 航, 牛建新. 核桃(L.) MicroRNA实时荧光定量RT-PCR内参基因的筛选[J]. 分子植物育种, 2019, 17(7): 2270-2278.
ZHOU L, QUAN S W, MA L, XU H, NIU J X. Screening of MicroRNA reference genes for Walnut by real-time fluorescence quantitative RT-PCR[J]. Molecular Plant Breeding, 2019, 17(7): 2270-2278. (in Chinese)
[27] 刘伟灿, 王 骐, 周永刚, 邓 宇, 赵利旦, 王兴超, 靳 京, 董园园, 王 南, 王法微, 陈 欢, 李晓薇, 李海燕. 大豆干旱胁迫下miRNA与mRNA荧光定量PCR内参基因的筛选[J]. 西北农林科技大学学报(自然科学版), 2016, 44(2): 61-67.
LIU W C, WANG Q, ZHOU Y G, DENG Y, ZHAO L D, WANG X C, JIN J, DONG Y Y, WANG N, WANG F W, CHEN H, LI X W, LI H Y. Selection of reference genes for quantitative polymerase chain reaction of miRNA and mRNA in soybean under drought stress[J]. Journal of Northest A & F University (Nature Science Edition), 2016, 44(2): 61-67. (in Chinese)
[28] ZHANG L C, YANG X Q, YIN Y Y, WANG J X, WANG Y W. Identifcation and validation of miRNA reference genes in poplar under pathogen stress[J]. Molecular Biology Reports, 2021, 48: 3357-3366.
[29] LYU S H, YU Y, XU S R, XU S R, CAI W W, CHEN G X, CHEN J J, PAN D M, SHE W Q. Identification of appropriate reference genes for normalizing miRNA expression in citrus infected bysubsp.[J]. Genes(Basel), 2019, 11(1): 17.
[30] MACHADO R D, CHRISTOFF A P, LOSS-MORAIS G, MARGIS-PINHEIRO M, MARGIS R, KÖRBES A P. Comprehensive selection of reference genes for quantitative gene expression analysis during seed development in[J]. Plant Cell Reports, 2015, 34(7): 1139-1149.
Selection of miRNA Reference for Normalization of Quantitative Real-time PCR Analysis in the Bark of Rubber Tree (Muell. Arg.)
WU Shaohua, ZHANG Shixin, YANG Shuguang, TIAN Weimin*
Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences / Key Laboratory of Biology and Genetic Resources of Rubber Tree, Ministry of Agriculture and Rural Affairs / State Key Laboratory Breeding Base of Cultivation and Physiology for Tropical Crops, Haikou, Hainan 571101, China
Rubber tree is the main source of natural rubber worldwide. Natural rubber is synthesized and stored in laticifer, a tissue composed of laticifer cells. The laticifer cells in the trunk bark are directly associated with natural rubber production and differentiated from the fusiform initials of vascular cambia. As the number of laticifer rings is positively correlated with rubber yield, the differentiation of laticifer from vascular cambia is a major theoretical subject faced to natural rubber industry. Plant miRNAs are a class of small noncoding RNAs about 20‒24 nucleotides in length and play a pivotal regulatory role in development, cell differentiation and adversity stress by mediating the the gene silencing. Quantitative Real-time PCR (qPCR) is widely used in the quantitative analysis of the miRNA expression levels, and the selection of appropriate internal reference of miRNA is crucial for accurating the miRNA expression levels. In the present study, the expression stability of three non-coding RNAs (,,) and 6 mature miRNAs (,,,,,) were evaluated on the basis of coronatine-induced secondary laticifer differentiation in the bark of rubber trees using poly (A)-tailed qPCR. According to the analysis of geNorm and NormFinder,andwere the top two stable miRNAs andwas the least stable gene in response to coronatine in the cambium tissue of rubber trees. The results showed thatandcould serve as qPCR reference miRNA to analyze the miRNA expression pattern in COR-induced secondary laticifer differentiation. This study will provide a good basis for identification of the differentially expressed miRNAs related to secondary laticifer differentiation.
Muell. Arg.; laticifer differentiation; miRNA; coronatine; reference gene
S794.1
A
10.3969/j.issn.1000-2561.2022.11.001
2022-03-28;
2022-06-23
海南省基础与应用基础研究计划(自然科学领域)高层次人才资助项目(No. 2019RC334);国家天然橡胶产业技术体系育种技术与方法岗位科学家项目(No. CARS-33-YZ1)。
吴绍华(1983—),男,博士,副研究员,研究方向:橡胶树分子生物学。*通信作者(Correponding author):田维敏(TIAN Weimin),E-mail:wmtian@163.com。

