二氢黄酮醇-4-还原酶在花青素合成中的功能及调控研究进展
李亚丽,李 欣,肖 婕,李瑞玲,杨华丽,孙 勃,汤浩茹
(四川农业大学 园艺学院,成都 611130)
植物色素主要有三大类:花青素、类胡萝卜素和生物碱类色素,其中花青素是一种天然的水溶性色素,它可使植物组织和器官呈现出不同的颜色,成为衡量果树果实品质和观赏植物观赏价值的重要指标之一。同时,花青素不仅可以帮助植物抵御生物胁迫和非生物胁迫,包括保护植物免受病原菌侵染、抗紫外辐射和清除体内多余的活性氧[1],而且还具有良好的药理作用,可以抗氧化、抗衰老、防止癌症和心血管疾病等。植物中主要存在6种花青素,分别是天竺葵素(pelargonidin)、矢车菊素 (cyanidin)、飞燕草素(delphindin)、芍药素(peonidin)、矮牵牛素(petunidin)及锦葵素(malvidin)。花青素是类黄酮途径的重要代谢产物,经苯丙氨酸裂解酶(PAL)、肉桂酸-4-羟化酶(C4H)、4香豆酰辅酶A连接酶(4CL)、查尔酮合成酶(CHS)、查尔酮异构酶(CHI)、黄烷酮-3-羟化酶(F3H)、类黄酮3′ 羟化酶(F3′H)、类黄酮3′5′ 羟化酶(F3′5′H)、DFR、花青素合成酶(ANS)等一系列酶催化合成[2](图1),其中CHS、CHI和F3H三种酶均处于类黄酮途径的上游,对于花青素和黄酮醇的合成都十分关键,因而研究较多且功能已相对明确。有研究发现,位于该途径中下游的DFR蛋白对花青素的合成也至关重要,它不仅能引导类黄酮途径流向花青素合成方向[4],而且更重要的是它在一定程度上决定着花青素的种类与含量。DFR可催化二氢山柰酚(dihydrokaempferol,DHK)、二氢槲皮素(dihydroquercetin,DHQ)、二氢杨梅素(dihydromyricetin,DHM)、柚皮素(naringenin)和圣草酚(eriodictyol)生成不同的花青素前体,再经ANS催化生成不同的花青素使植物组织或器官所呈现出不同的颜色,有研究指出麝香兰(Muscari)中的DFR以催化DHM为主,使花表现为蓝色[5],而草莓(Fragaria×ananassa)果实中的DFR主要催化DHK,使其果实表现为红色[6]。此外,DFR的功能丧失会使植物的组织或器官颜色变淡或呈无色[7-9]。因而,大多数改变植物器官颜色的基因工程主要围绕DFR进行[10]。
1 DFR的生物学功能
1.1 DFR的特征
DFR一般以单基因或小基因家族的形式出现在植物中,拟南芥(Arabidopsis)、葡萄(Vitisvinifera)和番茄(Solanumlycopersicum)等植物基因组中仅有1个DFR基因[11],而矮牵牛(Petunia)、玉米(Zeamays)、非洲菊(Gerbera)、草莓和芜菁(Brassicarapa)等植物中存在2个甚至多个DFR基因[6, 12-15]。一些双子叶植物如拟南芥、矮牵牛和芜菁的DFR基因均含有6个外显子和5个内含子,且它们内含子的位置相同[11]。部分具有DFR小基因家族的植物如矮牵牛和百脉根(Lotuscorniculatus)分别有3和5个串联的DFR基因[16-17],而玉米中存在2个非串联的DFR基因[11]。大部分植物DFR编码300~400个氨基酸的蛋白,但也有少数编码的氨基酸数量不在此范围内,如铜绿微囊藻DFR仅有72个氨基酸,而龙须藻DFR拥有634个氨基酸。DFR编码的蛋白存在至少2个典型的结构域,其中1个是能与NADPH结合的结构域,该结构域具有1个高度保守的NADP结合模体TGXXGXX(X代表任意氨基酸残基)。另一个是在DFR编码蛋白的132~157位氨基酸附近有1个由26个氨基酸组成的底物结合区。软件预测的DFR具有短链脱氢酶(short-chain dehydrogenase/reductase,SDR)超家族的所有几何特征,包括保守的YXXXK模体,略有差异的NADP结合模体和1个具有催化活性的丝氨酸。通过在大肠杆菌中异源表达葡萄VvDFR发现该晶体含有15个α螺旋,2个短310螺旋和12个β折叠[18]。
1.2 DFR的功能
DFR的底物比较宽泛,它可分别催化DHK、DHQ和DHM分别生成无色天竺葵素(leucopelargonidin)、无色矢车菊素(leucocyanidin)和无色飞燕草素(leucodelphinidin),随后经ANS催化生成天竺葵素、矢车菊素和飞燕草素(图1)。一些DFR可以同时催化DHK、DHQ和DHM,但对3种底物的催化效率不同[19]。有的DFR只能催化某种特定的二氢黄酮醇,如草莓中DFR1只能催化DHK,而DFR2可催化DHM和DHQ[6]。因DFR催化的底物不同使植物出现不同的颜色,如矮牵牛中的DFR可催化DHQ和DHM,使其花出现蓝色,因不能催化DHK,而使矮牵牛不能产生砖红色的花[20-21],但Meyer等[22]将玉米DFR转入突变为白花的矮牵牛中却产生了砖红色矮牵牛花。因此,DFR对底物的选择是决定着植物中花色苷的种类的重要因素并在很大程度上决定花色苷的比例。而使植物最终呈不同的颜色。此外,有研究发现DFR还可作为黄烷酮4-还原酶(flavanone 4-reductase,FNR),催化柚皮素和圣草酚生成黄烷-4-醇(flavan-4-ols),进而在其他酶的作用下形成3-脱氧花青素(3-deoxyanthocyanidin),这不仅使植物呈现出橙红色(图1),而且3-脱氧花青素及其中间代谢产物五羟基黄酮(luteoforol)还可作为防御物质抵御真菌和细菌侵害[23-25]。
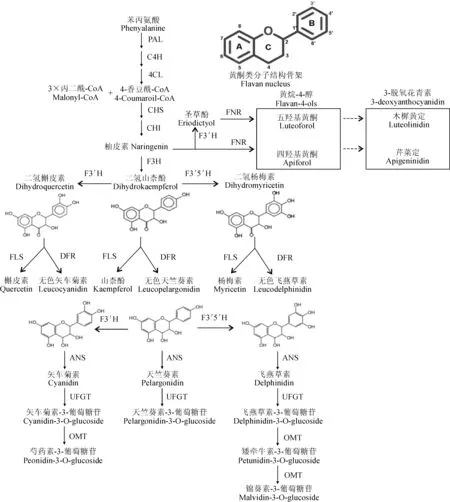
PAL. 苯丙氨酸裂解酶;C4H. 肉桂酸-4-羟化酶;4CL. 4-香豆酰辅酶A连接酶; F3′H. 类黄酮-3′-羟化酶;F3′5′H. 类黄酮-3′5′-羟化酶;FLS. 黄酮醇合成酶;ANS. 花青素合成酶;UFGT. 类黄酮葡萄糖苷转移酶; OMT. O-甲基转移酶图1 花青素的生物合成途径(改自Casanal A[3])PAL. Phenylalanine ammonia lyase;C4H. Cinnamate-4-hydroxylase;4CL. 4-coumaroyl:CoA-ligase;F3′H. Flavanone 3′ hydrolase;F3′5′H. Flavonoid 3′, 5′-hydroxylase;FLS. Flavonol synthase;ANS. Anthocyanidin synthase;UFGT. UDPGlucose-flavoniod glucosytransterase; OMT. O-methyl transferaseFig.1 Anthocyanins biosynthetic pathway(modified from Casanal A[3])
1.3 DFR的作用机制
目前,以葡萄DFR的作用机制最为清楚,而其他植物的DFR作用机制研究相对较少,葡萄VvDFR的结构显示其催化机制与SDR家族蛋白高度一致:NADP+与DFR通过氢键和静电相互作用结合,第163位络氨酸具有催化作用,第128位丝氨酸稳定底物,第167位赖氨酸通过与烟酰胺核糖形成氢键来降低第163位络氨酸的酸度系数而促进质子间的转运[18]。DHK、DHM和DHQ在B-环分别有1、2、3个羟基,其羟基化模式是DFR底物选择性的基础。Johnson等[10]通过对非洲菊的研究首次提出了DFR的底物结合区,并发现其中的第134位氨基酸种类是决定所催化底物的关键。有研究发现葡萄VvDFR的底物结合区有一个结合二氢黄酮醇B-环的口袋,其第133位天冬酰胺(与非洲菊第134位氨基酸对应)与底物B-环3′、4′ 位置的羟基形成氢键[18],进一步证实了DFR第133位氨基酸对其底物选择的重要作用。有研究指出矮牵牛PhDFR第133位氨基酸为天冬氨酸,可催化DHQ和DHM,但不能有效催化DHK[10]。如果将该位点同为天冬氨酸的舞春花(Calibrachoa×hybrida)CaDFR转入矮牵牛后,即可催化DHK产生天竺葵素[26],这可能是由于其他氨基酸产生的位阻效应阻碍了矮牵牛PhDFR与DHK结合所致。同样,天使花(Angeloniasalicariifolia)AngDFR的第12位丝氨酸和第26位甘氨酸以及大豆GmDFR2的第39位精氨酸突变后它们的花色均变浅[27-28],这也可能是突变后的氨基酸产生的位阻效应造成的。此外,虽然DFR的第133位氨基酸为天冬酰胺时可催化DHK和DHQ[29-30],但其催化效率只有50%和66%,如果将DFR第133位天冬酰胺突变为天冬氨酸,尽管它对DHK和DHQ的催化效率都有所降低,但仍然偏向于催化DHQ,这可能是第133位氨基酸不能单独决定DFR的底物选择性[18]。
1.4 DFR的系统进化
基因重复是蛋白进化出新功能的源泉。目前被大众熟知的进化机制有2种,分别是NEO-F(neofunctionalization )模型和EAC模型(escape from adaptive conflict)。NEO-F模型指基因复制后其中1个拷贝产生了新的功能,EAC则指基因复制后的2个拷贝都产生了新的功能且其原始功能依然存在[31]。Des Marais和Rausher[32]认为DFR基因重复的适应性变化为EAC模型,并根据此模型推测DFR最初可催化二氢黄酮醇、柚皮素和圣草酚,且其催化二氢黄酮醇的功能受到限制,在进化过程中,催化柚皮素和圣草酚的功能逐渐退化,而催化二氢黄酮醇的功能被释放出来。不同DFR拷贝在进化过程中功能出现差异,黄芩(Scutellaria)中存在2个DFR拷贝,通过分析其遗传进化,发现不平衡的正选择压力导致其不同拷贝的进化速度不同,致使其调控元件的数量、密码子偏好性和氨基酸等不同以致于DFR功能出现差异[33]。我们通过在NCBI搜集不同物种的DFR蛋白序列,利用MEGA4构建系统发育树(图2),该发育树可分为A、B、C3个分枝,A分枝物种均为单子叶植物,而B、C分枝为双子叶植物,与其他DFR进化树结果一致[11],说明DFR的趋异可能发生在单子叶与双子叶的趋异之后。我们的研究发现,在红蓝光下差异表达的草莓FaDFR3与草莓FaDFR1、FaDFR2的遗传距离相差甚远,FaDFR3在图2中C分枝上,该分枝上的DFR少有研究,功能尚不明确,但该枝上梨(Pyruspyrafolia)的DFR基因响应UV-B光[34],这些基因是否与非生物胁迫相关有待证实。
2 DFR的调控
2.1 环境和激素对DFR的调控
2.1.1环境因子对DFR的调控许多植物在非生物胁迫下会合成花青素以保护机体免受损伤,这个过程往往伴随着DFR表达量的升高。光可以从不同层面来影响花青素的含量,包括光质(红蓝光、紫外光)、光量、光周期、光的方向[35]。近年来光对花青素含量影响的研究层出不穷,强光、蓝光、紫外光均能诱导DFR表达以增加花青素的含量[36-39]。植物体内至少存在4种光受体:光敏色素,感受红光和远红光;隐花色素,感受蓝光和近紫外光;向光素,感受蓝光;UV-B受体,感受较短波长的紫外光[40]。这些受体接收光信号后将其传递给下游信号传导组分,如COP1,COP1可直接或间接调控MYB类转录因子,而MYB可以调节花青素合成结构基因(DFR等)的表达从而影响花青素的含量[41]。温度是影响花青素合成的又一关键因素,研究表明低温诱导花青素合成,而高温抑制花青素合成[42]。低温环境下MdMYB10、MdbHLH3/33、SlAH(bHLH)等转录因子表达增多,可促进DFR等结构基因的表达以提高花青素的含量[43-44]。然而也存在一些DFR受低温诱导但与花青素合成关系不大,芜菁中克隆得到的12个BrDFR中,BrDFR2、4、8、9受低温诱导并与花青素积累相关,而BrDFR1、3、5、6、10均受低温和冻害诱导且与花青素的积累量无关[12]。此外,土壤微生物、干旱、盐度和一定浓度的氮、磷与钙等也可以诱导花青素合成基因DFR的表达[45-50]。
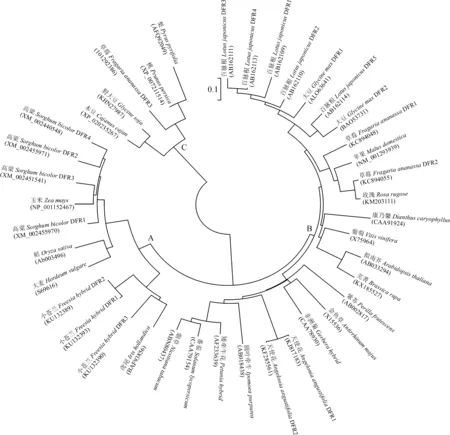
A~C代表3个不同的进化枝:A.单子叶植物;B和C.双子叶植物图2 DFR的系统进化树A-C are three different clades: A. monocotyledonous; B and C. dicotyledonousFig.2 The phylogenetic tree of DFR
2.1.2激素对DFR的调控激素对花青素的合成具有重要的作用。有研究发现,去掉草莓绿果期的瘦果可以促进DFR的表达,而对去瘦果的草莓果实喷施NAA后DFR的表达受到抑制[51]。草莓、葡萄、甜樱桃经ABA处理后花青素含量增多[52-54],而红肉苹果组培苗经ABA处理后花青素含量降低[55],与花青素合成相关的基因如DFR、MYB10等表达量减少,这种差异可能是由于苹果是呼吸跃变型果实,草莓、葡萄和樱桃(Prunusavium)属于非呼吸跃变型果实,而ABA在非呼吸跃变型果实的成熟中发挥着重要的作用,同时可以促进成熟相关的花青素合成。大部分文献表明脱落酸(ABA)、茉莉酸甲酯(JA)、细胞分裂素(CK)、油菜素内酯(BR)对花青素的合成有促进作用,且BR可以促进JA和CK诱导的花青素合成,而赤霉素(GA)和乙烯则抑制花青素的合成[56-59]。激素对花青素的调控主要是通过调控各种转录因子的活性,再由转录因子调控花青素合成后期结构基因(DFR等)的表达从而实现对花青素的调控[55,58]。
2.2 转录因子对DFR的调控
转录因子又称反式作用因子,是一类可直接或间接与目的基因启动子区域的顺式作用元件发生特异性结合,并对基因的转录起调节作用的蛋白。研究发现MYB、bHLH与WD40这3类转录因子形成的三元复合体MBW与黄酮类物质的合成密切相关[60]。MBW可活化花青素合成结构基因的启动子,从而促进花青素的合成。近年来,一些研究通过遗传、生物化学和分子手段证明了DFR是MBW复合体的靶基因之一[61]。MYB类转录因子对花青素的调控研究较多,苦荞FtMYB1和FtMYB2以及葡萄VvMYBA2等MYB可以促进DFR的表达,抑制FLS的表达[62-63],而草莓FaMYB5、淫羊藿(Epimediumsagittatum)EsMYBF1则抑制DFR的表达[64-65],箭叶淫羊藿EsMYBA1、甜樱桃PacMYBA、百合(Liliumspp.)LhMYB12已通过双荧光素酶检测分析发现其可活化DFR的启动子[66-68]。拟南芥中的光敏色素互作因子PIF4与PIF5(bHLH)可以抑制DFR的启动子活性从而负调控花青素的合成[69],而苹果(Malusdomestica)MdbHLH3可与DFR的启动子结合并促进它的表达[70]。紫罗兰(Matthiolaincana)TTG(WD40)的一个核苷酸发生替换导致氨基酸改变后,其DFR不表达,且花表现为白色[71]。此外,SPL9和NAC家族的ANAC0329等转录因子也可调控DFR的表达,且最近发现的miR156-SPL9-DFR通路还可以帮助植物抵御非生物胁迫[72-73]。
2.3 部分结构基因对DFR的影响
2.3.1F3′H和F3′5′H影响DFR功能的发挥F3′H可将DHK转化为DHQ,而F3′5′H可将DHK转化为DHM或将DHQ转化为DHM[74],从而限制DFR功能的发挥[75]。康乃馨(Dianthuscaryophyllus)、君子兰(Cliviaminiata)和玫瑰(Rosarugosa)均无蓝紫色花是因为它们缺乏F3′5′H活性而无法生成DHM所致[76];相反,蓝色三色堇(Violatricolor)花瓣中蓝色正是由于F3′5′H、DFR和ANS这3个基因的高表达的结果[77]。野生草莓(Fragariavesca)果实全红时期矢车菊素与天竺葵素的比值达0.51,远高于栽培草莓果实全红时期的比值(0.05),这是因为野草莓中F3′H的表达量比栽培草莓中的F3′H高,催化DHK生成了更多的DHQ,使矢车菊素所占的比例升高[6]。当缺少F3′H和F3′5′H活性的淡红色矮牵牛转入玫瑰DFR后,其花瓣和花药都变成粉橙色,因为F3′H和F3′5′H酶活缺失,导致DHQ和DHM无法产生,所以DFR只能催化DHK产生橙色的天竺葵素衍生物[78]。非洲菊‘passion’可产生矢车菊素和天竺葵素这2类花青素,当施用抑制F3′H活性的四环唑后,DHQ的浓度降低,天竺葵素衍生物含量增多[79],这已成为生产上改变非洲菊花色的重要手段。近年来有学者发现F3′H和F3′5′H不仅可以转化不同的二氢黄酮醇,还能转化不同的无色花色素,从而成为花青素合成新途径[80](图1)。
2.3.2FLS与DFR竞争底物FLS和DFR作用的底物相同,FLS可催化DHK, DHQ和DHM生成黄酮醇类物质山柰酚、槲皮素和杨梅素(图1)。研究表明,在白色矮牵牛花中过表达DFR,花变粉红色,而引入FLS的反义链抑制FLS活性后白色矮牵牛花也变为粉红色[81]。近年来,DFR和FLS的平衡关系影响花青素和黄酮醇的含量在多种植物中得到了验证。Luo等[82]选取了7个不同花色的植物(玫瑰、桃花、矮牵牛花)探讨其呈现出红花和白花的原因,最终发现DFR和FLS的不平衡表达导致了这样的花色差异。同样对白花麝香兰和蓝花麝香兰的转录组数据进行分析,发现白花麝香兰中DFR表达很低,而杨梅素和山柰酚含量显著升高,所以白花麝香兰蓝色缺失的主要原因是DFR表达降低导致黄酮醇类物质增多[5]。这样竞争底物的关系在蔬菜和果树中同样存在,将大豆GmMYB12B2转入拟南芥中,DFR表达水平降低,而FLS表达明显升高,黄酮醇类物质含量增多[83],而沉默紫色土豆(IpomoeabatatasLam.)的IbDFR后黄酮醇也大量增加[83]。苹果(Malusspp.)的果实和叶片中过表达McDFR和沉默McFLS都能使其花青素含量升高,红色加深,且过表达McFLS和沉默McDFR均能使果实和叶片红色变浅[84]。DFR和FLS对底物的竞争已成为调控许多植物呈色的关键方法。
3 展 望
近年来,随着分子生物学技术的发展以及花青素代谢途径研究的深入,DFR的结构与功能得到了进一步阐释,然而仍有许多问题尚不明确。首先,一些植物存在DFR基因家族,其家族不同成员的组织特异性不同,对各种胁迫的响应也不相同,表明其不同成员的功能存在差异,如高粱(Sorghumbicolor)中SbDFR1调控花青素的积累,而SbDFR3则控制3-脱氧花青素的生成[25],因而DFR基因家族的植物其不同成员的功能以及它们之间的关系需要进一步研究。其次,尽管已经证实一些转录因子可以活化DFR的启动子,但其顺式作用元件并不清楚,而当今ChIP等技术的高速发展为此研究提供了可能,且不同转录因子对DFR的调控不同,研究特定转录因子与DFR的关系可为今后改变植物组织或器官的颜色提供新的思路。此外, DFR蛋白的3D结构研究甚少,因而其对DFR底物选择性的机理仍不清楚,这可能是今后研究的一个方向。
参考文献:
[1]CHEN Q, YU H, TANG H,etal. Identification and expression analysis of genes involved in anthocyanin and proanthocyanidin biosynthesis in the fruit of blackberry[J].ScientiaHorticulturae, 2012, 141:61-68.
[2]JAAKOLA L. New insights into the regulation of anthocyanin biosynthesis in fruits[J].TrendsinPlantScience, 2013,18(9):477-483.
[3]CASANAL A, ZANDER U, MUNOZ C,etal. The strawberry pathogenesis-related 10 (PR-10) Fra a proteins control flavonoid biosynthesis by binding to metabolic intermediates[J].TheJournalofBiologicalChemistry, 2013,288(49):35 322-35 332.
[4]LIM S H, YOU M K, KIM D-H,etal. RNAi-mediated suppression of dihydroflavonol 4-reductase in tobacco allows fine-tuning of flower color and flux through the flavonoid biosynthetic pathway[J].PlantPhysiologyandBiochemistry, 2016, 109:482-490.
[5]LOU Q, LIU Y, QI Y,etal. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower colour in grape hyacinth[J].JournalofExperimentalBotany, 2014,65(12):3 157-3 164.
[6]MIOSIC S, THILL J, MILOSEVIC M,etal. Dihydroflavonol 4-reductase genes encode enzymes with contrasting substrate specificity and show divergent gene expression profiles inFragariaspecies[J].PLoSOne, 2014,9(11):e112707.
[7]KIM S, PARK J Y, YANG T J. Characterization of three active transposable elements recently inserted in three independent DFR-A alleles and one high-copy DNA transposon isolated from the Pink allele of the ANS gene in onion (AlliumcepaL.)[J].MolecularGeneticsandGenomics, 2015,290(3):1 027-1 037.
[8]ZHANG J, WENDELL D L, VAZIRI A,etal. The gene encoding dihydroflavonol 4-Reductase is a candidate for the anthocyaninless locus of rapid cyclingBrassicarapa(fast plants type)[J].PloSOne, 2016,11(8):e0161394.
[9]BAUMBACH J, PUDAKE R N, JOHNSON C,etal. Transposon Tagging of a Male-Sterility, female-sterility gene, St8, revealed that the meiotic MER3 DNA helicase activity Is essential for fertility in soybean[J].PLoSOne, 2016,11(3):e0150482.
[10]JOHNSON E T, RYU S, YI H,etal. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase[J].ThePlantJournal, 2001,25(3):325-333.
[11]INAGAKI Y, JOHZUKA-HISATOMI Y, MORI T,etal. Genomic organization of the genes encoding dihydroflavonol 4-reductase for flower pigmentation in the Japanese and common morning glories[J].Gene, 1999,226(2):181-188.
[12]AHMED N U, PARK J I, JUNG H J,etal. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress inBrassicarapa[J].Gene, 2014,550(1):46-55.
[13]HUITS H S, GERATS A G, KREIKE M M,etal. Genetic control of dihydroflavonol 4-reductase gene expression inPetuniahybrida[J].ThePlantJournal, 1994,6(3):295-310.
[14]SCHWARZ-SOMMER Z, SHEPHERD N, TACKE E,etal. Influence of transposable elements on the structure and function of the A1 gene ofZeamays[J].TheEMBOjournal, 1987,6(2):287.
[15]HELARIUTTA Y, ELOMAA P, KOTILAINEN M,etal. Cloning of cDNA coding for dihydroflavonol-4-reductase (DFR) and characterization of dfr expression in the corollas ofGerberahybridavar. Regina (Compositae)[J].PlantMolecularBiology, 1993,22(2):183-193.
[16]YOSHIDA K, IWASAKA R, SHIMADA N,etal. Transcriptional control of the dihydroflavonol 4-reductase multigene family inLotusjaponicus[J].JournalofPlantResearch, 2010,123(6):801-805.
[17]CHEN M, SANMIGUEL P, BENNETZEN J L Sequence organization and conservation in sh2/a1-homologous regions of sorghum and rice[J].Genetics, 1998,148(1):435-443.
[18]PETIT P, GRANIER T, D’ESTAINTOT B L,etal. Crystal structure of grape dihydroflavonol-4-reductase, a key enzyme in flavonoid biosynthesis[J].JournalofMolecularBiology, 2007,368(5):1 345-1 357.
[19]MARTENS S, TEERI T, FORKMANN G. Heterologous expression of dihydroflavonol-4-reductases from various plants[J].FEBSletters, 2002,531(3):453-458.
[20]FORKMANN G, RUHNAU B. Distinct substrate specificity of dihydroflavonol-4-reductase from flowers ofPetuniahybrida[J].ZeitschriftfürNaturforschungC, 1987,42(9-10):1 146-1 148.
[21]JOHNSON E T, YI H, SHIN B,etal. Cymbidium hybrida dihydroflavonol-4-reductase does not efficiently reduce dihydrokaempferol to produce orange pelargonidin-type anthocyanins[J].ThePlantJournal, 1999,19(1):81-85.
[22]MEYER P, HEIDMANN I, FORKMANN G,etal. A new petunia flower colour generated by transformation of a mutant with a maize gene[J].Nature, 1987,330(6 149): 677-678.
[23]LI H, QIU J, CHEN F,etal. Molecular characterization and expression analysis of dihydroflavonol 4-reductase (DFR) gene inSaussureamedusa[J].MolecularBiologyReports, 2012,39(3):2 991-2 999.
[24]FLACHOWSKY H, HALBWIRTH H, TREUTTER D,etal. Silencing of flavanone-3-hydroxylase in apple (MalusxdomesticaBorkh.) leads to accumulation of flavanones, but not to reduced fire blight susceptibility[J].PlantPhysiologyBiochemistry, 2012, 51:18-25.
[25]LIU H, DU Y, CHU H,etal. Molecular dissection of the pathogen-inducible 3-deoxyanthocyanidin biosynthesis pathway in sorghum[J].PlantandCellPhysiology, 2010,51(7):1 173-1 185.
[26]CHU Y X, CHEN H R, WU A Z,etal. Expression analysis of dihydroflavonol 4-reductase genes inPetuniahybrida[J].GeneticsandMolecularResearch, 2015,14(2):5 010-5 021.
[27]YAN F, DI S, RODAS F R,etal. Allelic variation of soybean flower color gene W4 encoding dihydroflavonol 4-reductase 2[J].BMCPlantBiology, 2014,14(1):58.
[28]GOSCH C, NAGESH K M, THILL J,etal. Isolation of dihydroflavonol 4-reductase cDNA clones fromAngeloniaxangustifoliaand heterologous expression as GST fusion protein inEscherichiacoli[J].PloSone, 2014,9(9):e107755.
[29]SHIMADA N, SASAKI R, SATO S,etal. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of theLotusjaponicusgenome[J].JournalofExperimentalBotany, 2005.56(419):2 573-2 585.
[30]XIE D Y, JACKSON L A, COOPER J D,etal. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase fromMedicagotruncatula[J].PlantPhysiology, 2004,134(3):979-994.
[31]STORZ J F. Genome evolution: gene duplication and the resolution of adaptive conflict[J].Heredity(Edinb), 2009,102(2):99-100.
[32]DES MARAIS D L,RAUSHER M D. Escape from adaptive conflict after duplication in an anthocyanin pathway gene[J].Nature, 2008,454(1 205):762-765.
[33]HUANG B H, CHEN Y W, HUANG C L,etal. Imbalanced positive selection maintains the functional divergence of duplicated dihydrokaempferol-4-reductase genes[J].ScientificReports, 2016, 6:39031.
[34]QIAN M, YU B, LI X,etal. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear,PyruspyrifoliaNakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/VIS conditions[J].PlantMolecularBiologyReporter, 2014,32(2):428-437.
[35]JIAO Y, LAU O S, DENG X W. Light-regulated transcriptional networks in higher plants[J].NatureReviewsGenetics, 2007,8(3):217-230.
[36]ZHANG K M, WANG J W, GUO M L,etal. Short-day signals are crucial for the induction of anthocyanin biosynthesis inBegoniasemperflorensunder low temperature condition[J].JournalofPlantPhysiology, 2016, 204:1-7.
[37]ALONSO R, BERLI F J, FONTANA A,etal. Malbec grape (VitisviniferaL.) responses to the environment: Berry phenolics as influenced by solar UV-B, water deficit and sprayed abscisic acid[J].PlantPhysiologyandBiochemistry, 2016, 109:84-90.
[38]JIANG M, REN L, LIAN H,etal. Novel insight into the mechanism underlying light-controlled anthocyanin accumulation in eggplant (SolanummelongenaL.)[J].PlantScience, 2016, 249:46-58.
[39]HONG Y, TANG X, HUANG H,etal. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum[J].BMCGenomics, 2015, 16:202.
[40]ZORATTI L, SARALA M, CARVALHO E,etal. Monochromatic light increases anthocyanin content during fruit development in bilberry[J].BMCPlantBiology, 2014, 14:377.
[41]MAIER A, SCHRADER A, KOKKELINK L,etal. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation inArabidopsis[J].ThePlantJournal, 2013,74(4):638-651.
[42]FERNANDES DE OLIVEIRA A, MERCENARO L, DEL CARO A,etal. Distinctive anthocyanin accumulation responses to temperature and natural UV radiation of two field-grownVitisviniferaL. cultivars[J].Molecules, 2015,20(2):2 061-2 080.
[43]QIU Z, WANG X, GAO J,etal. The tomato Hoffman’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that Is developmentally regulated and induced by low temperatures[J].PloSOne, 2016,11(3):e0151067.
[44]WANG N, ZHANG Z, JIANG S,etal. Synergistic effects of light and temperature on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malussieversiif. niedzwetzkyana)[J].PlantCell,TissueandOrganCulture(PCTOC), 2016,127(1):217-227.
[45]HILBERT G, SOYER J, MOLOT C,etal. Effects of nitrogen supply on must quality and anthocyanin accumulation in berries of cv. Merlot[J].Vitis-JournalofGrapevineResearch, 2015,42(2):69.
[47]XU W, PENG H, YANG T,etal. Effect of calcium on strawberry fruit flavonoid pathway gene expression and anthocyanin accumulation[J].PlantPhysiologyandBiochemistry, 2014, 82:289-298.
[48]IUCHI S, YAMAGUCHI-SHINOZAKI K, URAO T,etal. Novel drought-inducible genes in the highly drought-tolerant cowpea: cloning of cDNAs and analysis of the expression of the corresponding genes[J].PlantandCellPhysiology, 1996,37(8):1 073-1 082.
[49]ALI M B,MCNEAR D H. Induced transcriptional profiling of phenylpropanoid pathway genes increased flavonoid and lignin content inArabidopsisleaves in response to microbial products[J].BMCPlantBiology, 2014,14(1):84.
[50]KIM J, LEE W J, VU T T,etal. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance inBrassicanapusL[J].PlantCellReports, 2017,36(8):1 215-1 224.
[51]MOYANO E, PORTERO-ROBLES I, MEDINA-ESCOBAR N,etal. A fruit-specific putative dihydroflavonol 4-reductase gene is differentially expressed in strawberry during the ripening process[J].PlantPhysiology, 1998,117(2):711-716.
[52]SHEN X, ZHAO K, LIU L,etal. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (PrunusaviumL.)[J].PlantandCellPhysiology, 2014,55(5):862-880.
[53]BAN T, ISHIMARU M, KOBAYASHI S,etal. Abscisic acid and 2, 4-dichlorophenoxyacetic acid affect the expression of anthocyanin biosynthetic pathway genes in ‘Kyoho’grape berries[J].TheJournalofHorticulturalScienceandBiotechnology, 2003,78(4):586-589.
[54]LI D, LUO Z, MOU W,etal. ABA and UV-C effects on quality, antioxidant capacity and anthocyanin contents of strawberry fruit (FragariaananassaDuch.)[J].PostharvestBiologyandTechnology, 2014, 90:56-62.
[55]SUN J, WANG Y, CHEN X,etal. Effects of methyl jasmonate and abscisic acid on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malussieversiif. niedzwetzkyana)[J].PlantCell,TissueandOrganCulture(PCTOC), 2017,130(2):227-237.
[56]XU W, DUBOS C, LEPINIEC L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J].TrendsinPlantScience, 2015,20(3):176-185.
[57]ZHANG N, SUN Q, LI H,etal. Melatonin improved anthocyanin accumulation by regulating gene expressions and resulted in high reactive oxygen species scavenging capacity in cabbage[J].FrontiersinPlantScience, 2016, 7:197.
[58]PENG Z, HAN C, YUAN L,etal. Brassinosteroid enhances jasmonate-induced anthocyanin accumulation inArabidopsisseedlings[J].JournalofIntegrativePlantBiology, 2011,53(8):632-640.
[59]YUAN L B, PENG Z H, ZHI T T,etal. Brassinosteroid enhances cytokinin-induced anthocyanin biosynthesis inArabidopsisseedlings[J].BiologiaPlantarum, 2014,59(1):99-105.
[60]VALLARINO J G, OSORIO S, BOMBARELY A,etal. Central role of FaGAMYB in the transition of the strawberry receptacle from development to ripening[J].NewPhytologist, 2015,208(2):482-496.
[61]XU W, GRAIN D, BOBET S,etal. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets inArabidopsisseed[J].NewPhytologist, 2014,202(1):132-144.
[62]BAI Y C, LI C L, ZHANG J W,etal. Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis[J].PhysiologiaPlantarum, 2014,152(3):431-440.
[63]NIU T, GAO Z, ZHANG P,etal. MYBA2 gene involved in anthocyanin and flavonol biosynthesis pathways in grapevine[J].GeneticsandMolecularResearch, 2016,15(4):8 922.
[64]王玲, 汤浩茹, 王小蓉,etal. 利用 VIGS 技术研究草莓 FaMYB5 的功能[J]. 园艺学报, 2017,44(1):33-42.
WANG L, TANG H R, WANG X R,etal. Virs-induced gene silencing as a tool for FaMYB5 gene functional studies in strawberry[J].ActaHorticulturaeSinica, 2017,44(1):33-42.
[65]HUANG W, KHALDUN A B M, CHEN J,etal. A R2R3-MYB transcription factor regulates the flavonol biosynthetic pathway in a traditional Chinese Medicinal plant,Epimediumsagittatum[J].FrontiersinPlantScience, 2016, 7: e70778.
[66]HUANG W, SUN W, LV H,etal. A R2R3-MYB transcription factor fromEpimediumsagittatumregulates the flavonoid biosynthetic pathway[J].PLoSOne, 2013,8(8):e70778.
[67]RAVAGLIA D, ESPLEY R V, HENRY-KIRK R A,etal. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunuspersica) by a set of R2R3 MYB transcription factors[J].BMCPlantBiology, 2013,13(1):1.
[68]LAI Y-S, SHIMOYAMADA Y, NAKAYAMA M,etal. Pigment accumulation and transcription of LhMYB12 and anthocyanin biosynthesis genes during flower development in the Asiatic hybrid lily (Liliumspp.)[J].PlantScience, 2012, 193:136-147.
[69]LIU Z, ZHANG Y, WANG J,etal. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light inArabidopsisseedlings[J].PlantScience, 2015, 238:64-72.
[70]XIE X B, LI S, ZHANG R F,etal. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples[J].Plant,Cell&Environment, 2012,35(11):1 884-1 897.
[71]DRESSEL A,HEMLEBEN V. Transparent Testa Glabra 1 (TTG1) and TTG1-like genes inMatthiolaincanaR. Br. and related Brassicaceae and mutation in the WD-40 motif[J].PlantBiology, 2009,11(2):204-212.
[72]CUI L-G, SHAN J-X, SHI M,etal. ThemiR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants[J].ThePlantJournal, 2014,80(6):1 108-1 117.
[73]MAHMOOD K, XU Z, EL-KEREAMY A,etal. TheArabidopsistranscription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses[J].FrontierinPlantScience, 2016, 7:1 548.
[74]BRUGLIERA F, BARRI‐REWELL G, HOLTON T A,etal. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus ofPetuniahybrida[J].ThePlantJournal, 1999,19(4):441-451.
[75]CASTELLARIN S D, DI GASPERO G, MARCONI R,etal. Colour variation in red grapevines (VitisviniferaL.): genomic organisation, expression of flavonoid 3′-hydroxylase, flavonoid 3′, 5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin[J].BMCGenomics, 2006,7(1):12.
[76]TANAKA Y, TSUDA S, KUSUMI T. Metabolic engineering to modify flower color[J].PlantandCellPhysiology, 1998,39(11):1 119-1 126.
[77]LI Q, WANG J, SUN H-Y,etal. Flower color patterning in pansy (Viola×wittrockianaGams.) is caused by the differential expression of three genes from the anthocyanin pathway in acyanic and cyanic flower areas[J].PlantPhysiologyandBiochemistry, 2014, 84:134-141.
[78]TANAKA Y, FUKUI Y, FUKUCHI-MIZUTANI M,etal. Molecular cloning and characterization ofRosahybridadihydroflavonol 4-reductase gene[J].PlantandCellPhysiology, 1995,36(6):1 023-1 031.
[79]BASHANDY H, PIETIAINEN M, CARVALHO E,etal. Anthocyanin biosynthesis ingerberacultivar ‘Estelle’ and its acyanic sport ‘Ivory’[J].Planta, 2015,242(3):601-611.
[80]SCHWINN K, MIOSIC S, DAVIES K,etal. The B-ring hydroxylation pattern of anthocyanins can be determined through activity of the flavonoid 3′-hydroxylase on leucoanthocyanidins[J].Planta, 2014,240(5):1 003-1 010.
[81]DAVIES K M, SCHWINN K E, DEROLES S C,etal. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase[J].Euphytica, 2003,131(3):259-268.
[82]LUO P, NING G, WANG Z,etal. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants[J].FrontierinPlantScience, 2015, 6:1257.
[83]WANG H, FAN W, LI H,etal. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses[J].PLoSOne, 2013,8(11):e78484.
[84]TIAN J, HAN Z Y, ZHANG J,etal. The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples[J].ScientificReports, 2015, 5:12 228.
——矮牵牛

