Effects of nanofertilizer and nano-plant hormone on soil chemical properties and microbial community in two different soil types
John Lester Viscara PIDE ,Nolissa Delmo ORGANO ,Andre Freire CRUZ ,Lilia M.FERNANDO ,Lucille C.VILLEGAS ,Evelyn F.DELFIN ,Michelle Ann Magat CALUBAQUIB,Roselle Estera MADAYAG and Erlinda S.PATERNO
1Division of Soil Science,Agricultural Systems Institute,College of Agriculture and Food Science(CAFS),University of the Philippines Los Baños(UPLB),College,Laguna 4031(Philippines)
2Graduate School of Life and Environmental Sciences,Kyoto Prefectural University,1-5 Shimogamohangi-cho,Sakyo-ku,Kyoto 606-8522(Japan)
3Crop BiotechnologyDivision,Institute of Crop Science,CAFS,UPLB,College,Laguna 4031(Philippines)
4MicrobiologyDivision,Institute of Biological Sciences,College of Arts and Sciences,UPLB,College,Laguna 4031(Philippines)
5Institute of Plant Breeding,CAFS,UPLB,College,Laguna 4031(Philippines)
6Isabela State University Cabagan Campus,Isabela 3328(Philippines)
ABSTRACT Application of nanotechnology in agriculture has been expanded to improve crop production.The impact of nanomaterials(NMs)on factors that influence the survival and function of beneficial microorganisms is a less studied aspect that needs to be better understood.Only a few studies have assessed the effects of NMs on beneficial soil microorganisms.This study was conducted to assess the effects of nanofertilizer FertiGroe® N(FG-N)and nano-plant hormone HormoGroe® auxin(HG-A)on the chemical properties and microbial communities of two contrasting soils,Lipa clay loam(CL)and Sariaya sandy loam(SL),over a 35-d incubation period in the laboratory.Bacterial and fungal communities were evaluated using amplicon sequencing analysis within the 16S and internal transcribed spacer regions,respectively.The application of FG-N significantly decreased soil pH,but did not affect total N and available P for both soil types.A significant increase in exchangeable K was observed only in Lipa CL.The application of HG-A had no significant effect on soil chemical properties.Regarding the bacterial community after incubation,the relative abundances of Acidobacteriia,Chthonomonadetes,and Saccharimonadia decreased,whereas Acidimicrobiia,Chloroflexia,and Gemmatimonadetes increased with FG-N application in Lipa CL.The application of HG-A increased the relative abundance of Rubrobacteria,Chthonomonadetes,and Chloroflexia in Lipa CL.For the fungal community,FG-N application increased the relative abundance of Sordariomycetes,Agaricomycetes,and Eurotiomycetes,whereas Dothideomycetes and Mortierellomycetes decreased in Lipa CL after incubation.In Sariaya SL,FG-N application increased the relative abundance of Dothideomycetes,Eurotiomycetes,and Mortierellomycetes,but decreased that of Sordariomycetes and Agaricomycetes.Fungal classes observed in the control samples were not detected in the HG-A treatment,but were recovered after incubation in Lipa CL.The microbial diversity in both soil types showed slight changes with FG-N and HG-A application.Principal coordinate analysis illustrated the clustering of bacterial and fungal taxa between Lipa CL and Sariaya SL.Pearson correlation analysis showed that several bacterial and fungal communities were positively or negatively correlated with soil pH.The results suggest that FG-N can be safely used in crop production and HG-A may be used mainly for vegetative propagation.
Key Words: amplicon sequencing,bacterial community,crop production,fungal community,microbial diversity,nanomaterial,nanotechnology
INTRODUCTION
Nanotechnology deals with nanomaterials(NMs)working through manipulation,characterization,and manufacturing of materials at the nanometer scale ranging from 1 to 100 nm (Heet al.,2019).Over the past decade,the development of engineered NMs has resulted in a rapid expansion of the nanotechnology industry,owing to the unique physical,chemical,and biological properties of NMs at the nanoscale.Nanomaterials are increasingly used in numerous applications including food packaging,textiles,catalysts,biosensors,medical devices,and agriculture(Collinset al.,2012;Dimkpa and Bindraban,2018;Mahawar and Prasanna,2018;Raiet al.,2018).The practical applications of nanotechnology in agriculture and food industry have expanded across a wide spectrum in the development of nano-based sensors,nanocomposites,nanoremediation,nanopesticides,and nanofertilizers(Yataet al.,2018;Heet al.,2019).The development of nanopesticides,nanofertilizers,and nanoplant hormones has gained in popularity in recent years.
Engineered NMs in the form of nanofertilizers provide one or more required nutrients to plants and boost their growth and yield(Liu and Lal,2015;Adisaet al.,2019).Nanofertilizers have high surface area to volume ratio and sorption capacity,nanometer system,and distinctive properties,which render them more effective than conventional fertilizers (Kaundalet al.,2017).Nanofertilizers can enhance the use efficiency of both macro-and micronutrients through slow and controlled release,thereby reducing nutrient loss due to leaching,degradation,and volatilization and,perhaps,lowering the rates of fertilizer application(Manjunathaet al.,2016;Kaundalet al.,2017;Subramanian and Thirunavukkarasu,2017;Dimkpa and Bindraban,2018;Mahawar and Prasanna,2018).Plant hormones in nano form have the capacity to improve root and shoot length,biomass,and uptake efficiency compared to commercially available growth stimulants.
Upon recognition of the importance of NMs,a group of researchers from the University of the Philippines Los Baños(UPLB)developed the nanofertilizer FertiGroe®and the nano-plant hormone HormoGroe®(HG).FertiGroe®N,P,and K nanofertilizers are controlled-release fertilizers that improve nutrient uptake by plants and increase crop yield.HormoGroe®,on the other hand,is a nanoformulation of plant hormones derived from naturally occurring plant growth-promoting bacteria and is mainly produced by the encapsulation of nano-sized granules for controlled delivery.
Although the application of NMs offers some benefits to soil fertility,there is limited information concerning their effects on indigenous soil microflora.Soil microorganisms are of significant environmental importance,because they are responsible for various biogeochemical processes,such as nutrient cycling,waste degradation,and agricultural production,and have the ability to buffer environmental change through bioremediation(Ben-Mosheet al.,2013).Factors such as soil structure,pH,climate,water content,and biotic activity are among the parameters responsible for complex microbial diversity in soil (Rajendhran and Gunasekaran,2008).
The impact of NMs on factors that influence the survival and function of beneficial microorganisms is a less studied aspect that needs to be better understood.Only a few studies have assessed the effects of NMs on beneficial soil microorganisms(Juárez-Maldonadoet al.,2021;Khanet al.,2022).In addition,several studies have reported that variations in soil properties influence the bioavailability and bioreactivity of engineered NMs (Kaundalet al.,2017).Aside from being directly affected by NMs,soil properties and soil microbial communities also influence one another.Microbial composition affects soil chemical properties and the changes in soil chemistry can,in turn,influence the succession of microorganisms in the soil.In addition,the inherent composition of the soil may either enhance or mask the effects of NMs.
In this study,we determined the direct effects of nanofertilizer FertiGroe®N (FG-N) and nano-plant hormone HormoGroe®auxin(HG-A)on soil chemical properties and microbial community composition.We tested these NMs in two soil types,Lipa clay loam(CL)and Sariaya sandy loam(SL),with contrasting physicochemical properties.In addition,we performed a correlation analysis between the microbial composition and soil properties.
MATERIALS AND METHODS
Soil collection,preparation and analyses
Lipa CL samples were collected from soil surface(0—20 cm depth)of an experimental area in the Farmers Operation Division,UPLB(14°08′45.1′′N,121°15′43.5′′E).Sariaya SL samples were collected from Barangay Canda,Sariaya,Quezon,Philippines(13°55′18′′N,121°32′42′′E).These soil types were chosen because:1)they are the most common soil types used for agricultural production and 2)they have contrasting physicochemical properties and are,therefore,more suitable for comparison purposes.Prior to any treatment,the bulk samples were air-dried,pulverized,and passed through a 2-mm sieve.Subsamples were subsequently taken for chemical analyses.Soil pH was analyzed following the potentiometric method in a 1:2.5(weight:volume)soil/water suspension(PCARR,1980).Total N was analyzed using the micro-Kjeldahl method(Bremner and Mulvaney,1982).Available P was determined using the Bray P-2 extraction method with 0.1 mol L-1HCl and 0.03 mol L-1NH4F extractant(Bray and Kurtz,1945).The amount of P in the extract was quantified according to the method of Murphy and Riley(1962).Exchangeable K was extracted using the ammonium acetate method (PCARR,1980) and quantified using a MultiSkan Sky spectrophotometer (Thermo Fisher Scientific,Waltham,USA).Soil particle size distribution was determined using the hydrometer method(Bouyoucos,1962)and soil textural classes were determined based on the USDA soil texture classification triangle(Rowell,1994).
Incubation experiment
The effects of FG-N and HG-A on soil microbial community were evaluated through a laboratory incubation experiment conducted in the Soil Microbiology Laboratory,Division of Soil Science,Agricultural Systems Institute,College of Agriculture and Food Science,UPLB.Preliminary studies have indicated that these products do not have harmful effects on culturable bacteria and fungi(Basayet al.,2021a,b).The NMs used in the experiment were obtained from the National Institute of Molecular Biology and Biotechnology,UPLB.A 2 × 4 factorial experiment was conducted for both FG-N and HG-A.The first factor for FG-N was soil type(Lipa CL and Sariaya SL).The second factor was fertilization treatment,consisted of the following:control (without fertilizer),FG-N (150 kg N ha-1),urea(150 kg N ha-1),and zeolite-nanocarrier(Z-NC)(150 kg ha-1).For HG-A,the first factor was soil type(Lipa CL and Sariaya SL).The second factor was fertilization treatment,consisted of the control (without fertilizer),50 mg kg-1standard indole acetic acid(IAA),50 mg kg-1HG-A,and 50 mg kg-1HG carrier(phosphatidylcholine).Each treatment combination was replicated three times and arranged in a randomized complete block design.
Approximately 500 g soil sample was placed in a polypropylene bag and soil moisture content was adjusted to field capacity using sterile distilled water.The moist soil was pre-incubated for a week before the treatments were applied.The treated soil was incubated in the laboratory at room temperature ranging from 26 to 35°C for 35 d.Soil moisture content was maintained at field capacity throughout the incubation period by weighing the soil and adding sterile distilled water whenever necessary.Soil samples were collected at the beginning(day 1)and end(day 35)of the incubation period.Samples for DNA extraction(0.25 g)were aseptically collected,placed in 1.5-mL sterile Eppendorf tubes,and stored at-20°C.Samples for chemical analyses were air-dried,passed through a 2-mm sieve,and stored at room temperature.
Soil DNA extraction
Total genomic DNA from the soil samples was extracted in the Soil Microbiology Laboratory,UPLB using a Qiagen DNeasy Powersoil®DNA isolation kit(Qiagen,Carlsbad,USA)according to the manufacturer’s instructions.To obtain sufficient genomic DNA,the replicates from each treatment were pooled.The presence of DNA after the extraction was confirmed by agarose gel electrophoresis.The DNA quantity and quality were determined using a MultiSkan Sky microplate spectrophotometer(Thermo Fisher Scientific).
Amplicon sequencing of total genomic DNA
Amplicon polymerase chain reaction(PCR)and library construction of both 16S rDNA and internal transcribed spacer (ITS) genes in each sample were performed at the Pomology Laboratory,Kyoto Prefectural University,Japan.The PCR amplification and library construction for bacterial metagenome (16S rDNA) were conducted using primers targeting the V3—V4 regions as per the Fluidigm protocol(www.fluidigm.com).The PCR amplification was performed in a 20 μL mixture containing 10 μL KAPA Taq 2 ×ReadyMix buffer,2 μL genomic DNA,4 μL forward primer,and 4 μL reverse primer.The PCR conditions were as follows:95°C for 3 min,25 cycles of 95°C for 30 s,55°C for 30 s,and 72°C for 39 s,and a final elongation step at 72°C for 5 min.
The ITS gene sequences were amplified using primer pair ITS1/ITS2 and respective adapters(Table I),followed by index PCR for library construction(https://jp.illumina.com).Amplicon PCR for fungal metagenome was performed in a 20 μL mixture containing 10 μL KODFX buffer,1 μL genomic DNA,0.4 μL KOD FX Neo enzyme(Toyobo Co.,Osaka,Japan) (https:/www.toyobo-global.com/seihin/xr/lifescience/products/pcr_017.html),2 μL forward primer,2 μL reverse primer,4 μL dNTPs,and 0.6 μL sterile water.The PCR conditions were as follows:94°C for 2 min,30 cycles of 94°C for 15 s,55°C for 30 s,and 68°C for 1 min,and hold at 4°C.
All PCR products were cleaned using PROMEGA Wizard®SV Gel and PCR Clean-Up System(Madison,USA).The cleaned PCR products were subjected to index PCR(library construction) to attach dual indices and Illumina sequencing adapters using a Nextera XT index kit(Illumina Co.,San Diego,USA)(https://jp.illumina.com/techniques/sequencing/ngs-library-prep.html).The DNA libraries were validated,pooled,and subjected to amplicon sequencing using Illumina MiSeq 250 bp (Illumina Co.) at Genome Quebec in Montreal,Canada.
Sequence data analysis
The gene sequences for bioinformatics analysis were processed using the QIIME 2.0 pipeline(Bolyenet al.,2019),including decompression and re-assembly.The taxonomic base analysis for the 16S gene was accessed by 16S classifier provided in the QIIME 2.0 website (https://qiime2.org/),whereas ITS classifier was downloaded from Unite community(https://unite.ut.ee/).Software QIIME 2.0 was used to estimate the taxonomic metrics between communities to determine the relative abundance,operational taxonomic units (OTUs),and diversity (Shannon-Wiener index) of the microbes in soils.Principal coordinate analysis (PCoA) was performed for microbial community using R statistical software with respective packages.Sequences were deposited in the DNA Data Bank of Japan(DDBJ) (https://www.ddbj.nig.ac.jp) under accession number DRA012725.

TABLE IForward and reverse primers and adapters for 16S rDNA and internal transcribed spacer(ITS)
Statistical analysis for soil chemical properties
The chemical properties of the soil samples were subjected to the analysis of variance(ANOVA)using Statistical Tool for Agricultural Research(STAR 2.0.1).If significant,the treatment means were compared using the least significant difference(LSD)test at a 5%level of significance.The correlations between soil chemical properties and microbial community were determined using SPSS 19.0(IBM Corp.,Armonk,USA).
RESULTS AND DISCUSSION
Soil physicochemical properties
The Lipa soil series is classified as fine clayey,mixed,shallow,isohyperthermic Typic Eutrudepts,whereas the Sariaya soil series is classified as loamy,mixed,and isohyperthermic Cumulic Hapludolls(Caratinget al.,2014).These soil types were specifically selected for this study,because they are known to have contrasting properties.Preliminary soil analysis confirmed distinct differences in the physicochemical properties between the two soils(Table II).Lipa CL had higher amounts of silt and clay particles,with clay content five times higher than that of Sariaya SL.In contrast,the sand content of Sariaya SL was two times higher than that of Lipa CL.The results showed that both soils were moderately acidic(pH=5.40 and 5.80).The total N content of Lipa CL was two times higher,available P was three times higher,and exchangeable K was slightly higher than that of Sariaya SL.

TABLE IIInitial physicochemical properties of two soil types,Lipa clay loam(CL)and Sariaya sandy loam(SL)
Effects of FG-N and HG-A on chemical properties of Lipa CL and Sariaya SL
Statistical analysis revealed that soil type had a significant direct effect on the soil chemical properties measured in the experiment.The FG-N treatment had a direct effect on soil pH but did not directly influence total N,available P,and exchangeable K(Table SI,see Supplementary Material for Table SI).In contrast,the HG-A treatment directly affected available P and had no significant effect on soil pH,total N,and exchangeable K after the 35-d incubation(Table SII,see Supplementary Material for Table SII).Interaction effects were also observed between soil type and FG-N in terms of soil pH,total N,and exchangeable K.
Similar to the preliminary analysis,the chemical properties of Lipa CL were consistently higher than those of Saraiya SL after applications of FG-N and HG-A(Table III).Total N and exchangeable K in Lipa CL were almost twice those in Sariaya SL.Such difference in the magnitude of increase in total N and exchangeable K could be attributed to the higher sorption capacity of CL-textured soil due to its higher surface area and smaller particle size,and thereby greater retention of nutrients compared to the SL-textured soil.

TABLE IIIMeans of soil pH,total N,available P,and exchangeable K in two soil types,Lipa clay loam (CL) and Sariaya sandy loam (SL),after application of nanofertilizer FertiGroe® N(FG-N)and nano-plant hormone HormoGroe®auxin(HG-A)for 35 d
The FG-N and Z-NC treatments significantly lowered the pH of both soil types compared with the control(Table SI).Despite having a lower initial pH(pH 5.02)than Z-NC-treated Lipa CL(pH 5.29),the pH of Sariaya SL still decreased to 4.76 after application of Z-NC(Table SI).This suggests that the material present in Z-NC can promote further reduction in soil pH regardless of soil type.Interestingly,the total N content of FG-N-treated soils was not significantly different from that of the control,even though the recommended rate of 150 kg N ha-1was used (Table SI).A previous study reported that a N nanofertilizer synthesized using zeolite released higher amounts of available N than the control and conventional fertilizer during a 30-din vitroincubation(Rajoneeet al.,2016).Since the release of N from FGN has not been studied yet,we can only assume that a large amount of N from FG-N had already been released and eventually volatilized before the end of the incubation period,resulting in total N almost similar to that of the control.A significant interaction effect between treatment and soil type was observed in the urea treatment.The urea-treated Lipa CL significantly lower total N compared with the control.In contrast,urea-treated Sariaya SL had a slightly higher total N than the control(Table SI).The observed reduction in total N in urea-treated soil supports the idea that volatilization might be high in Lipa CL,leading to a lower total N content despite the application of urea.
HormoGroe®application significantly increased soil available P(Table SII),which can be attributed to the phospholipid wall material used to encapsulate auxin.This is an advantage because it can also serve as a source of P.Although plant growth regulators are usually applied as foliar spray,they may reach the soil during periods of high precipitation.Nevertheless,this does not seem to have any negative effects on soil chemical composition.
Effects of FG-N and HG-A on bacterial communityof Lipa CL and Sariaya SL
The number of bacterial sequences per sample ranged from 35 774 to 46 118,with an average of 40 906.8 and a total of 409 068 good-quality 16S rDNA gene sequences from all samples.The relative abundance of bacterial groups was uniform among all samples on day 1.On day 35,differences were observed between soil type and nanomaterial treatment(Fig.1).
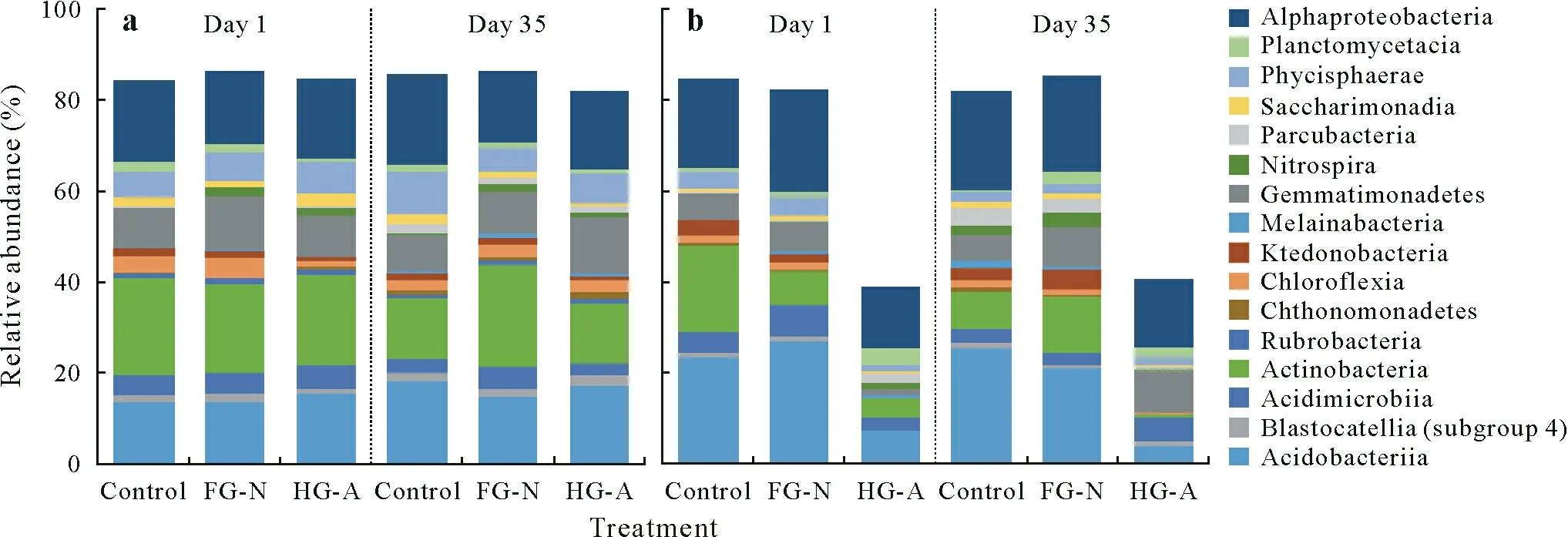
Fig.1 Distributions of bacterial classes in two soil types,Lipa clay loam(a)and Sariaya sandy loam(b),with application of nanofertilizer FertiGroe® N(FG-N)and nano-plant hormone HormoGroe® auxin(HG-A)on days 1 and 35.
The assignment of bacterial sequences at the class level revealed the influence of soil type on the composition of bacterial community(Fig.1).Lipa CL was strongly dominated by Actinobacteria with the relative abundance of approximately 15%.In contrast,Alphaproteobacteria was the most abundant bacterial class in Sariaya SL,with relative abundance of approximately 18%.Bacterial groups that were commonly dominant in both soil types included Alphaproteobacteria,Actinobacteria,Acidobacteriia,Gemmatimonadetes,Phycisphaerae,and Acidimicrobiia.For both soil types,the relative abundance of Acidobacteriia,Parcubacteria,and Alphaproteobacteria increased after 35 d of incubation(DOI).In contrast,the relative abundance of Actinobacteria decreased from 21.45%to 13.43%in Lipa CL and from 19.08%to 8.10%in Sariaya SL.
The effect of soil type on microbial community remained evident in nanomaterial-treated soils(Fig.1).The relative abundance of Actinobacteria was significantly higher in Lipa CL than in Sariaya SL after application of FG-N for 35 d.The relative abundance of Acidobacteriia,Chthonomonadetes,Saccharimonadia,Phycisphaerae,and Alphaproteobacteria declined with FG-N application in Lipa CL.In contrast,the relative abundance of Acidimicrobiia,Chloroflexia,Gemmatimonadetes,and Nitrospira increased with FG-N application in Lipa CL.
The relative abundance of bacterial groups Acidimicrobiia,Chloroflexia,and Melainabacteria decreased in Sariaya SL treated with FG-N;however,the relative abundance of Actinobacteria,Ktedonobacteria,Gemmatimonadetes,Nitrospira,and Planctomycetacia were significantly enriched.The application of HG-A increased the relative abundance of Rubrobacteria,Chthonomonadetes,Chloroflexia,Gemmatimonadetes,and Nitrospira in Lipa CL and the relative abundance of Acidimicrobiia,Gemmatimonadetes,and Planctomycetacia in Sariaya SL(Fig.1).It should be noted that HG-A is not recommended for direct application in soil because plant growth-promoting hormones are usually applied as a foliar fertilizer.
These results confirm the effect of soil type on microbial community composition.The bacterial community structure appears to be shaped by soil texture.The effect of soil type on bacterial community has been well documented.Proteobacteria,Firmicutes,Actinobacteria,Acidobacteria,and Bacteriodetes were observed to dominate clay loam soil in the plant-associated bacterial community(Citlaliet al.,2018).In another experiment,the dominant bacterial phyla in sandy loam soil were Proteobacteria,Actinobacteria,Acidobacteria,Bacteroidetes,Chloroflexi,Gemmatimonadetes,Cyanobacteria,and Firmicutes(Rajoneeet al.,2016).
The interaction effect between soil type and nanomaterial treatment was also evident.The responsive bacterial groups included Actinobacteria (Actinobacteria),Acidimicrobiia(Acidobacteria),Chloroflexia(Chloroflexi),Gemmatimonadetes (Gemmatimonadetes),and Nitrospira (Nitrospirae).These bacterial groups could be useful in monitoring possible changes as indicator organisms when NMs are applied to the soil.
Nitrogen is by far the most important element in fertilizers and is a critical nutrient source in agriculture.For instance,N in the form of ammonium significantly affects the soil bacterial community composition (Myroldet al.,2014).Only a few studies have been conducted on the effects of NMs on beneficial soil microorganisms.Thomaset al.(2016)observed that the application of 80 mg kg-1of biosynthesized nano-N significantly increased bacterial population of Nitrosomonas,Nitrobacter,Azotobacter,and actinomycetes and improved the activities of enzymes such as alkaline phosphatase,acid phosphatase,esterase,and dehydrogenase.
Acidobacteria play crucial environmental roles,as indicated by their roles in key C,N,and S biogeochemical cycles(Kalamet al.,2020).In addition,Acidobacteria have been identified as one of the key bacterial taxa in soil,emphasizing their importance in C cycling and their role in the decomposition of soil organic matter(Banerjeeet al.,2016).Actinobacteria are Gram-positive bacteria that can break down cellulose and chitin,which are the primary sources of soil nutrients,and are well-known for their filamentous growth.Soil texture influences the abundance of Actinobacteria and its sublevel taxa(Myroldet al.,2014).Bacteria belonging to the phylum Gemmatimonadetes comprise approximately 2%of soil bacterial communities,are typically seen in 16S rRNA in the natural environment,and have been identified as one of the top nine phyla found in soils(DeBruynet al.,2011).Conversely,little is known about the ecological functions of this phylum in soil and the factors that influence its abundance(Chenet al.,2016).Chloroflexi are phototrophic Gram-negative bacteria(Mohammadi and Prasanna,2003),and their growth and reproduction in terrestrial habitats are not dependent on the supply of soil nutrients.
Effects of FG-N and HG-A on bacterial diversityof Lipa CL and Sariaya SL
In the FG-N treatment,the number of observed OTUs was higher in Sariaya SL than in Lipa CL (Fig.S1,see Supplementary Material for Fig.S1).The analysis based on the Shannon-Wiener index showed very little to no distinct changes in bacterial diversity in both soil types after application of FG-N.The effect of soil type on bacterial diversity was evident in the HG-A treatment(Fig.S2,see Supplementary Material for Fig.S2).The Shannon-Wiener index slightly decreased in Sariaya SL from 6.48 on day 1 to 6.37 on day 35,whereas it increased from 6.27 to 7.24 in Lipa CL(Fig.S2).The PCoA at the family level revealed distinct clustering between Lipa CL and Sariaya SL(Fig.2).

Fig.2 Principal coordinate(PC)analysis plot of operational taxonomic units in bacterial communities at family level in two soil types,Lipa clay loam(CL)and Sariaya sandy loam(SL),with application of nanofertilizer FertiGroe®N(FG-N)and nano-plant hormone HormoGroe®auxin(HG-A).LC-F=Lipa CL in control for FG-N;LF=Lipa CL in FG-N;SC-F=Sariaya SL in control for FG-N;SF=Sariaya SL in FG-N;LC-H=Lipa CL in control for HG-A;LH=Lipa CL in HG-A;SC-H=Sariaya SL in control for HG-A;SH=Sariaya SL in HG-A.
These results demonstrate that the difference in soil type had strong effects on the soil bacterial community composition and could either mask or amplify the influence of nanofertilizers.In relation to the difference in soil types ranging from coarse sand to silty clay loam(pH 4.6—5.7),Chauet al.(2011)reported that bacterial species richness was higher in coarser-textured soils,which provide suitable microhabitat that fosters higher richness.In a controlled inoculation experiment on sandy loam soil,a positive correlation between bacterial richness and aggregate stability was observed in bare soil and uninoculated treatments(Martinet al.,2012).This corroborates the reports that soil types shape the diversity of soil microbial community structure in clay,loam,and loamy-sand soils(Obayomiet al.,2021)and sandy and clay loam soils(Griffithset al.,2008).
Correlation analysis between soil properties and soil bacterial community
The relative abundance of Acidobacteriaceae,Acidimicrobiaceae,Ktedonobacteraceae,Peptostreptococcaceae,Nitrospiraceae,and Xanthobacteraceae were negatively correlated with soil pH,whereas those of Rubrobacteriaceae and Hyphomicrobiaceae were positively related with soil pH and total N (Table IV).The relative abundance of Streptomycetaceae was positively correlated with total N,exchangeable K,and dehydrogenase activity,but negatively correlated with available P.Both Ktedonobacteraceae and Peptostreptococcaceae had a positive relationship with available P.In contrast,Pearson correlation analysis showed that urease activity had no significant influence on the bacterial community.
Factors such as soil pH,texture,structure,climate,water content,and enzyme activity are among the parameters that result in complex microbial diversity.Soil pH,along with other physicochemical parameters,has a significant influence on the community composition of soil bacteria(Zhanget al.,2016;Lladóet al.,2018).Patterns of community composition in silt loam soil showed a significant correlation with soil physicochemical properties,such as pH,N,P,and K(Degruneet al.,2016).Xiaet al.(2020)found that soil pH was both positively and negatively correlated with Acidobacteria and its sublevel taxa.
Effects of FG-N and HG-A on fungal communityin Lipa CL and Sariaya SL
The number of sequences per sample ranged from 42 231 to 75 456,with an average of 52 956.6 and a total of 529 566 good-quality ITS gene sequences from all samples.Amplicon sequencing revealed the influence of soil type on fungal community in the two soils.Sordariomycetes(Ascomycota),Mortierellomycetes(Mortierellomycota),Agaricomycetes(Basidiomycota),and Dothideomycetes(Ascomycota)were the dominant fungal groups in both soil types throughout the incubation period(Fig.3).Sordariomycetes,the most abundant class among the samples,accounted for an average of 47%of the total population.After 35 DOI,certain fungal groups behaved differently between the two soil types.The relative abundance of fungal groups Dothideomycetes,Eurotiomycetes,and Mortierellomycetes increased in Lipa CL but decreased in Sariaya SL.

Fig.3 Distributions of fungal classes in two soil types,Lipa clay loam(a)and Sariaya sandy loam(b),with application of nanofertilizer FertiGroe® N(FG-N)and nano-plant hormone HormoGroe® auxin(HG-A)on days 1 and 35.
The application of FG-N increased the relative abundance of fungal classes Sordariomycetes,Agaricomycetes,and Eurotiomycetes in Lipa CL after 35 DOI.In contrast,the proportions of Dothideomycetes and Mortierellomycetes decreased(Fig.3).The application of HG-A initially reduced the fungal composition of Lipa CL on day 1.The relative abundance of Sordariomycetes in the HG-A treatment was significantly lower than that in the control.The majority of the fungal classes observed in the control were not detected in the HG-A treatment.After 35 DOI,the fungal population in the HG-A treatment recovered,as the majority of the fungal groups increased in abundance except Dothideomycetes and Mortierellomycetes(Fig.3).
In Sariaya SL,FG-N application increased the relative abundance of Dothideomycetes,Eurotiomycetes,and Mortierellomycetes,but decreased the relative abundance of Sordariomycetes and Agaricomycetes.While reduced fungal abundance was observed in HG-A-treated Lipa CL,HG-A-treated Sariaya SL showed a different trend.The relative abundance of fungal classes Dothideomycetes,Eurotiomycetes,and Rhizophlyctidomycetes was higher in the HG-A treatment on both days 1 and 35.Sordariomycetes initially showed a negative response to HG-A treatment,but their relative abundance increased on day 35.
The influence of soil texture on the members of Ascomycota and Basidiomycota was negatively correlated with high sand content and positively correlated with silt/claycontent (Xiaet al.,2020).In the study of Guerreroet al.(2019),Ascomycetes (Ascomycota) and Basidiomycetes(Basidiomycota)were isolated from Lipa CL.Ascomycota and Basidiomycota were found to be the dominant fungal phyla in sandy loam soils(Panget al.,2021).

TABLE IVPearson correlation coefficients between soil properties and relative abundance of bacteria at family level
Ascomycota,a fungal phylum with the most species,accounted for nearly 75%of all known fungal species.Ascomycota and Basidiomycota are part of the subkingdom Dikarya(Lee Taylor and Sinsabaugh,2015).Additionally,both phyla have the highest genetic and ecological diversity,which is aided by the expression of a range of different polysaccharidedegrading enzymes.Ascomycota,also known as cup fungi,are a key factor in the C and N cycles;they improve soil stability for erosion control and seasonal plant biomass decomposition particularly during the early stages of decomposition,and directly interact with plants as endophytes(Lee Taylor and Sinsabaugh,2015;Challacombeet al.,2019).The phylum Basidiomycota or club fungi comprise a few groups of fungi that form beneficial symbiotic relationships with plants,but a significant number of the mechanisms by which these fungi absorb nutrients have yet to be identified (Jinet al.,2019;Martha-Pazet al.,2019).Mortierellomycota,a saprophytic phylum,have the ability to grow quickly and a preference for dissolved C sources(Qinet al.,2014).
Effects of FG-N and HG-A on fungal diversity in Lipa CL and Sariaya SL
The number of observed OTUs decreased from 373.4 to 339.4 and from 400.0 to 299.2 in Lipa CL and Sariaya SL,respectively,following the FG-N application for 35 d(Fig.S3,see Supplementary Material for Fig.S3).The Shannon-Wiener index slightly decreased upon FG-N application.Lipa CL and Sariaya SL treated with HG-A showed an increase in the number of observed OTUs compared with the control(Fig.S4,see Supplementary Material for Fig.S4).The Shannon-Wiener index increased from 5.02 to 5.84 in Lipa CL and from 4.99 to 6.55 in Sariaya SL in the HG-A treatment after 35 DOI(Fig.S4).
The PCoA plot of taxonomical abundance showed that fungal community composition clustered in Lipa CL and Sariaya SL(Fig.4).The separation between the control and nanofertilizer treatments was observed in Lipa CL,which was slightly different in Sariaya SL.Within cluster,the effect of nanofertilizer treatments was more significant for the fungal community,particularly in Lipa CL.Fungi are more morphologically complex than bacteria but still physiologically similar.This suggests that,although fungi have lower diversity,the effect was more specific to nanofertilizer treatments in different soil types.
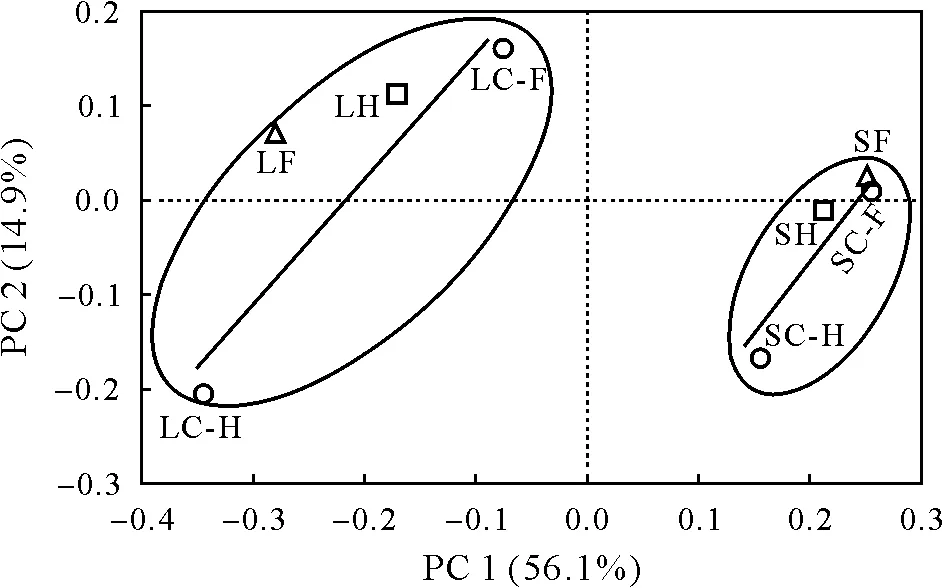
Fig.4 Principal coordinate(PC)analysis of operational taxonomic units in fungal community in two soil types,Lipa clay loam(CL)and Sariaya sandy loam(SL),with application of nanofertilizer FertiGroe® N(FG-N)and nano-plant hormone HormoGroe® auxin(HG-A).LC-F=Lipa CL in control for FG-N;LF=Lipa CL in FG-N;SC-F=Sariaya SL in control for FG-N;SF=Sariaya SL in FG-N;LC-H=Lipa CL in control for HG-A;LH=Lipa CL in HG-A;SC-H=Sariaya SL in control for HG-A;SH=Sariaya SL in HG-A.
Correlation analysis between soil properties and soil fungal community
Several fungi were positively or negatively correlated with soil pH at the family level(Table V).In the fungal community,the abundance of Cladosporiaceae and Nectriaceae was positively correlated with soil pH and total N.The abundance of Sordariaceae was positively correlated with soil pH,exchangeable K,and dehydrogenase and urease activities,whereas the abundance of Trichocomaceae,Nectriaceae,and Glomeraceae was negatively correlated with available P.The abundance of Morosphaeriaceae,Sporormiaceae,Herpotrichiellaceae,and Clavicipitaceae was negatively correlated with dehydrogenase activity and exchangeable K.Fertilization provides available nutrients to soil microorganisms,which enhance soil nutrient inputviarhizosphere deposition (Sunet al.,2016).Most fungi require aerobic conditions and organic matter as sources of C and energy for their growth and reproduction;therefore,competition within the fungal population may be more intense(Xiaet al.,2020).It remains unclear how and to what extent changes inmicrobial diversity affect soil ecosystem functions and stability(Degruneet al.,2016).Because of the unknown health and environmental risks associated with NMs,the improbabilities regarding possible interactions with environmental factors have made identifying these risks with nanoscale structure difficult(Heet al.,2019).
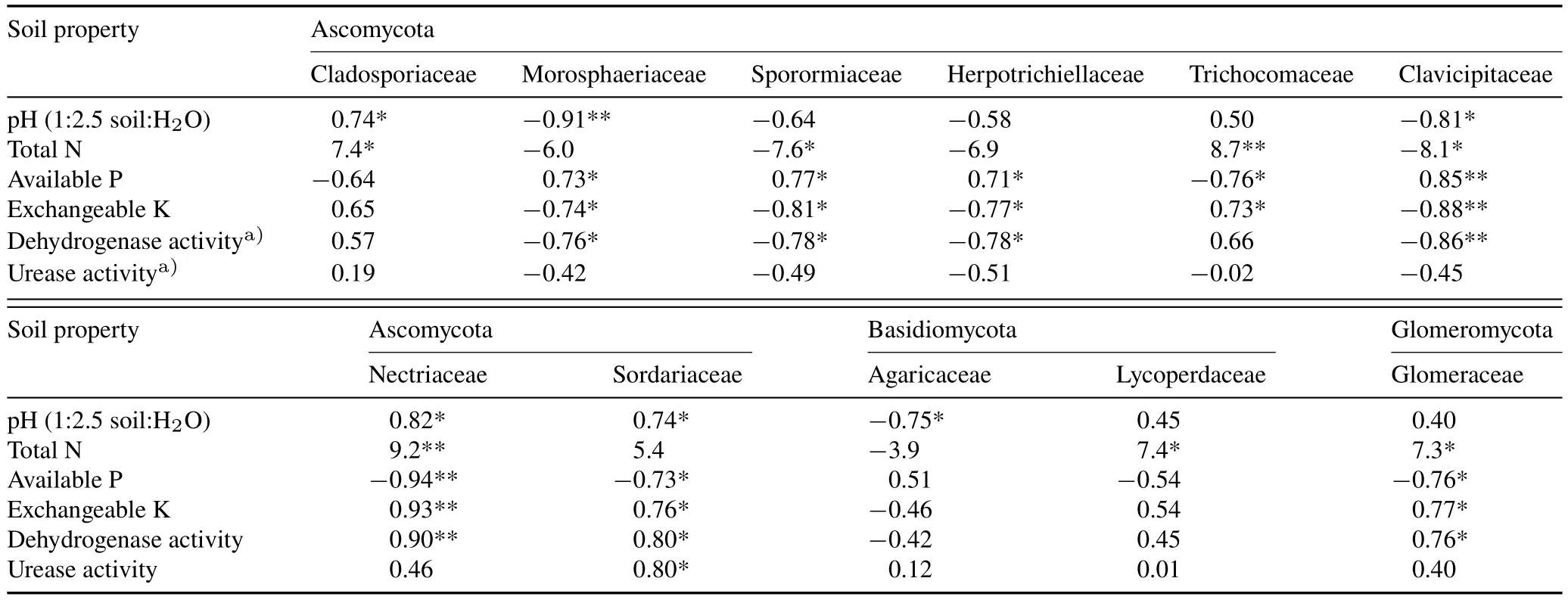
TABLE VPearson correlation coefficients between soil properties and relative abundance of fungi at family level
CONCLUSIONS
Amplicon sequencing analysis revealed the effects of soil type and interaction with NMs on the bacterial and fungal community composition.Lipa CL was strongly dominated by Actinobacteria,and Sariaya SL by Alphaproteobacteria.Sordariomycetes were most abundant among the samples.The FG-N and HG-A applications were significantly correlated with the abundances of Chloroflexia,Gemmatimonadetes,Nitrospira,Acidimicrobiia,and Actinobacteria in bacteria and Sordariomycetes,Agaricomycetes,Dothideomycetes,Eurotiomycetes,and Mortierellomycetes in fungi.These groups can be used as indicator organisms to monitor possible changes in organisms after NM application to the soil.Nanofertilizer may be safely used for crop production,whereas the application of HG-A directly to the soil should be limited and may be used mainly for vegetative propagation.
In this study,the effect of soil type on bacterial diversity was evident in the HG-A treatment after 35 DOI.Fungal OTUs and Shannon-Wiener index in both Lipa CL and Sariaya SL decreased 35 d after FG-N application and increased 35 d after HG-A application.Clustering of bacterial and fungal community composition at family level with nanofertilizer treatments showed a distinct grouping between Lipa CL and Sariaya SL.Streptomycetaceae,Ktedonobacteraceae,and Peptostreptococcaceae were correlated with soil pH,available P,and total N.At family level,Sordariaceae,Trichocomaceae,Nectriaceae,and Glomeraceae were associated with soil chemical properties.
Because the effects of nanomaterials on soil microbial community may differ under laboratory conditions and the natural environment,there is a need to conduct experiments in field for longer incubation periods.Furthermore,research should include responsive bacterial and fungal groups that could be later used as indicator organisms in NM-amended soil.The results of this study provide a basis for policy recommendations regarding the acceptability of NMs.
ACKNOWLEDGEMENTS
This study was funded by the Department of Science and Technology (DOST),Philippine Council for Agriculture,Aquatic and Natural Resources Research and Development(No.N9-102-2A)and the DOST,Accelerated Science and Technology Human Resource Development Program of Philippines.We thank Ms.Shaira Mhel Joy Granada and Ms.Ma.Czet Fulleros from the Division of Soil Science,Philippines,Ms.Hosne A.Dilzahan from Kyoto Prefectural University,Japan,and Mr.Carlito P.Basay Jr.from University of Southern Mindanao,Philippines for their assistance with soil sampling and analysis.
SUPPLEMENTARY MATERIAL
Supplementary material for this article can be found in the online version.
- Pedosphere的其它文章
- Carbon farming by recarbonization of agroecosystems
- Root exclusion methods for partitioning of soil respiration:Review and methodological considerations
- Utilization of lignocellulosic plant residues for compost formation and its role in improving soil fertility
- A critical review of microbially induced carbonate precipitation for soil stabilization:The global experiences and future prospective
- Hand-feel soil texture observations to evaluate the accuracy of digital soil maps for local prediction of soil particle size distribution:A casestudy in Central France
- Influence of soil physicochemical properties,particle size fractions and mineralogy on the leaching potentials of arsenic and antimony in abandoned mine soils

