Main environmental drivers of abundance,diversity and community structure of comammox Nitrospira in paddy soils
Fuyun GAO ,Yaying LI ,Haoxin FAN ,Jiantao XUE and Huaiying YAO,,*
1Key Laboratory of Urban Environment and Health,Institute of Urban Environment,Chinese Academy of Sciences,Xiamen 361021(China)
2Zhejiang Key Laboratory of Urban Environmental Processes and Pollution Control,CAS Haixi Industrial TechnologyInnovation Center in Beilun,Ningbo 315800(China)
3University of Chinese Academy of Sciences,Beijing 100049(China)
4Research Center for Environmental Ecologyand Engineering,School of Environmental Ecologyand Biological Engineering,Wuhan Institute of Technology,Wuhan 430073(China)
ABSTRACT The recently discovered complete ammonia oxidizers comammox Nitrospira contain clades A and B that can establish an independent one-step nitrification process;however,little is known about their environmental drivers or habitat distributions in agricultural soils.Previous studies on comammox Nitrospira in paddy soils have mainly focused on small-scale samples,and there is a lack of multisite research on comammox Nitrospira in paddy soils.In this study,we conducted a survey of 36 paddy soils to understand the community structure,abundance,and diversity of comammox Nitrospira and the degree to which they are affected by environmental factors at a large scale.Comammox Nitrospira were found to be widely distributed among the paddy soils.The abundance of comammox Nitrospira clade A was mostly lower than that of clade B,whereas its diversity was mostly higher than that of clade B.Correlation analysis showed that multiple factors affected(P <0.05)the abundance of comammox Nitrospira,including soil pH,organic matter,total carbon,and total nitrogen,latitude,mean annual temperature,and mean annual precipitation.Moreover,there was a clear relationship between the comammox Nitrospira community and habitat,indicating that some amplicon sequence variants(ASVs)had a unique dominant position in specific habitats.Phylogenetic analysis showed that the ASVs of comammox Nitrospira clade A clustered with the known sequences in the paddy soils and were significantly different from the known sequences in other habitats,which may be related to the unique paddy field habitat.In contrast,comammox Nitrospira clade B showed no clear habitat dependence.These results support the wide distribution and high abundance of comammox Nitrospira in paddy soils and provide novel insights into nitrogen cycling and nutrient management in agricultural ecosystems.
Key Words: ammonia-oxidizing archaea,ammonia-oxidizing bacteria,amplicon sequence variant,complete ammonia oxidizer,environmental factor,habitat,nitrification,nitrogen cycling
INTRODUCTION
Paddy soils are widely distributed in China,and paddy ecosystems are an important component of global agriculture(Huet al.,2015),covering more than 158 million hectares of arable land(Timilsinaet al.,2020).The drying-wetting stage of paddy soils provides a unique environment for microorganisms(Ishiiet al.,2011).Ammonia oxidation plays a vital role in the global nitrogen cycle.As the first rate-limiting step,it is executed by ammonia-oxidizing archaea(AOA)and ammonia-oxidizing bacteria(AOB).However,recent evidence has confirmed that complete ammonia oxidizers(comammoxNitrospira)can independently perform complete ammonia oxidation(Daimset al.,2015;Van Kesselet al.,2015).According to the difference in ammonia monooxygenase,comammoxNitrospirais separated into clades A and B(Daimset al.,2015;Van Kesselet al.,2015;Palomoet al.,2018).
The abundance of comammoxNitrospiraclades A and B responded differently to various environmental factors that have been studied;however,the response of the community of ammonia oxidizers to environmental factors and habitat remains unknown(Liuet al.,2018;Liu S Fet al.,2020;Shiet al.,2020).Previous studies have indicated that comammoxNitrospiradisplay high yield,high substrate affinity,relatively low maximum oxidation rate,and slow growth under highly oligotrophic conditions(Van Kesselet al.,2015;Kitset al.,2017).These results support the early hypothesis that slow growth in ammonia-depleted biofilms,soils,or sediments represents the fundamental niche of comammoxNitrospira(Costaet al.,2006;Kitset al.,2017).The abundance of comammoxNitrospirais lower than the abundance of AOA and/or AOB in agricultural and forest soils(Liet al.,2020;Shiet al.,2020;Wanget al.,2020)or approximately equal to the abundance of AOB in certain forest soils(Shiet al.,2020).In contrast,comammoxNitrospiraclade A has been shown to outnumber clade B in alkaline paddy soils(Wanget al.,2018)and AOA in acidic forest soils(Hu and He,2017).ComammoxNitrospirapreferred a weakly alkaline environment,because the optimal pH of key Nconverting enzymes(e.g.,hydroxylamine dehydrogenase and ammonia monooxygenase)is approximately 7.0—8.0(Blumet al.,2018).Studies have examined the influence of various environmental factors on each ammonia oxidizer community,including soil temperature(Liu Z Bet al.,2020),soil salinity(Liu Z Bet al.,2020),fertilization(Wanget al.,2018),and geographic factors(Liuet al.,2019;Shiet al.,2020;Zhanget al.,2020).However,this is a relatively new field of study,and further research into the relationship between the comammoxNitrospiracommunity and various environmental factors is required.
Paddy ecosystems vary in time and space,and the dryingwetting stage of paddy soils provides a unique environment for microorganisms(Ishiiet al.,2011).The Northeast Plain,the Yangtze River basin,and the southeast coast are the three most important rice-producing regions of China.Therefore,paddy fields cover a wide range of climatic zones(Houet al.,2020).Drying-wetting alternation in paddy soils causes regular fluctuations in soil redox potential (Ahmedet al.,2019)and frequent aerobic-anoxic alternations.Although comammoxNitrospirahave been studied in engineering systems,the driving factors for nitrification and the largescale distribution of comammoxNitrospirain paddy soils remain poorly understood.Deciphering the effects of various environmental factors on the abundance,diversity,and community of comammoxNitrospirain soil is essential.Therefore,we collected soils from 36 paddy fields across China to determine soil characteristics and molecular properties and to study the main environmental drivers affecting the abundance,diversity,and community of comammoxNitrospiraclades A and B.Here,we applied real-time quantitative polymerase chain reaction (qPCR) and high-throughput sequencing combined with QIIME2(Bolyenet al.,2019)to comprehensively analyze the communities of comammoxNitrospiraclades A and B.Through this study,we aimed to understand the following:i)the distribution,abundance,diversity,and community characteristics of comammoxNitrospirain Chinese paddy soils;ii)the environmental driving factors of comammoxNitrospira;and iii)the phylogenetic differences in the comammoxNitrospiracommunity between paddy soils and other known habitats such as engineering systems,sediments,and forest soils.The results of this study could be applied to enhance the management strategy of nitrogen cycle in paddy soils as a result of improved understanding of the main environmental drivers affecting the functions of comammoxNitrospiraclades A and B.
MATERIALS AND METHODS
Sample collection and soil properties
In accordance with data from the National Bureau of Statistics of China(http://www.stats.gov.cn/),the sampling points set up in this study fully considered the dominant rice planting areas,planting years (>15 years),and planting scale to ensure the typicality of paddy soils.Thirty-six sites were selected for soil sampling from 12 provinces in Northeast,East,Central,South,and Southwest China.From July to September 2018,soil was sampled at rice harvest,with three replicates per site.Approximately 2 kg of soil comprising eight soil cores(5 cm in diameter)was randomly collected from the top 0—20 cm soil layer across the entire field at each site,mixed well,and divided into three composite samples.The samples were placed into sampling bags and transported on ice to the laboratory.Each sample was again divided into three parts.One part was air-dried for chemical analysis,another part was stored at 4°C for biochemical analysis,and the third part was freeze-dried for DNA extraction.Latitude and longitude,climate type,mean annual temperature(MAT),and mean annual precipitation(MAP)of these 36 sites are shown in Tables SI and SII(see Supplementary Material for Tables SI and SII).
Soil pH value was measured in a soil to water ratio of 1:2.5(weight:volume)using a pH meter(Thermo Fisher Scientific,Waltham,USA).Total carbon(TC)and total nitrogen(TN)contents were determined by combustion decomposition using an elemental analyzer(Elementar Analysensysteme GmbH,Langenselbold,Germany).Microbial biomass carbon(MBC)and microbial biomass nitrogen(MBN)were calculated from the differences between samples fumigated with chloroform(CHCl3)and those unfumigated and then extracted with 0.5 mol L-1potassium sulfate(K2SO4),as described by Menget al.(2020).Soil ammoniumand nitratewere extracted by shaking with a potassium chloride (KCl) solution for 1 h.The ratio of soil to KCl was 1:10(weight:volume)and the KCl concentration in the final solution was 0.02 mol L-1.Soil organic matter(SOM)content was measured using potassium permanganate(K2Cr2O7)oxidation-volumetric analysis,as described by Menget al.(2021).
DNA extraction and qPCR
Genomic DNA was extracted from each sample(500 mg of soil) using a FastDNA spin kit for soil (MP Biomedicals,Cleveland,USA) according to the manufacturer’s instructions.Before conducting downstream analysis,the concentration and quality of DNA were detected with a NanoDrop ND-2000 UV-Vis spectrophotometer(NanoDrop Technologies,Wilmington,USA).
We used qPCR to quantify the functional gene(amoA)involved in ammonium oxidation in comammoxNitrospira.Equimolar mixtures of six pairs of primers for comammoxNitrospiraclades A and B,namely comaA-244f_a—f/comaA-659r_a—f and comaB-244f_a—f/comaB-659r_a—f,were used(Pjevacet al.,2017).Real-time qPCR was performed with SYBR®Premix Ex Taq II(Promega,Madison,USA)in a Roche LightCycler 480 instrument(Roche Molecular Systems,Basel,Switzerland).Each reaction was executed in a 20 μL volume containing approximately 5 ng of DNA,0.25 μmol L-1of each primer,and 10 μL of SYBR®Premix Ex Taq II.Primers and thermal cycling conditions are listed in Table SIII(see Supplementary Material for Table SIII).Target band was separated by electrophoresis on 2%agarose gel and then cut out.After purification,the target band was loaded into PGM-T Easy Vector system(Tian Gen,Beijing,China)to build the clone library.Positive clones were selected for sequencing and extracting the plasmids ofamoA.The qPCR standard curves were generated by 10×gradient dilution of extractedamoA-positive plasmids.The negative control contained the same amount of water with DNA as the template.The efficiency of all qPCR experiments was greater than 88%and the error was less than 0.200.
Functional gene(amoA)sequencing
The comammoxNitrospiracommunities in paddy soils were analyzed using Illumina sequencing of comammoxNitrospiraclades A and B functional geneamoA.Highthroughput paired-end Illumina MiSeq sequencing(PE 300)was performed at Majorbio(Shanghai,China).After sequencing,Illumina MiSeq fastq reads of the comammoxNitrospiraclades A and BamoAgene were analyzed using DADA2 plugin in QIIME2(https://qiime2.org/).The 8 bp barcode was collated and distributed to each sample and all reads<375 bp were cut offand discarded.The trimmed sequences were Chimera-detected and deleted using the vsearch algorithm,which can supposedly represent the real biological sequences present in the sample,known as amplicon sequence variants(ASVs).The sampling depth was 1 312 for comammoxNitrospiraclade A and 1 291 for comammoxNitrospiraclade B.Clustering of high-quality sequences into ASVs was performed with vsearch pipeline at almost 100%similarity level.The neighbor-joining method was used to build a phylogenetic tree based on top 1(>1%genus)representative sequences.
The original sequences of the comammoxNitrospira amoAgene have been submitted to GenBank under the accession Nos.SRR13149211—SRR13149411.
Statistical analysis
SPSS 22 for Windows(IBM Corp.,Armonk,USA)was used to calculate Spearman correlation coefficient,which was used to explain the significant correlation between pH,MAT,MAP,geochemical properties,and the abundance of comammoxNitrospiraclades A and B.Redundancy analysis (RDA) was conducted to identify environmental factors driving the differences between comammoxNitrospiraclades A and B.Prior to RDA,species-sample data were subjected to a detrended correspondence analysis to estimate gradient lengths;for values<3,the RDA analysis was initiated.The representative sequences obtained in this study were compared with the reference sequences defined in the National Center for Biotechnology Information (NCBI) database using BioEdit,and a phylogenetic tree was constructed using MEGA-X,with 1 000 bootstrap replicates (Kumaret al.,2018).A phylogenetic tree was built by first translating nucleotide sequences into amino acid sequences.The sequence data were analyzed using QIIME2,and vsearch pipeline was used to cluster high-quality sequences into amplicon variants at a 100%similarity level.Aggregated boosted tree(ABT)models first converted the ASV table into a beta diversity distance and then converted all environmental factors into a matrix of differences between samples.The matrices were converted into a single column of data and cross-validation was performed.All other statistical tests were conducted using ggplot2,vegan,dismo,gbm,and reshape2 packages in R software(v.4.0.2).
RESULTS
Distribution and abundance of comammox Nitrospira clades A and B
The abundance and distribution of comammoxNitrospiraclades A and B were compared among the paddy soils by determining the abundance ofamoAgene(Fig.1).TheamoAgene abundance of clades A and B ranged from 8.27×105to 3.14×108copies g-1dry soil and from 4.94×106to 9.67×108copies g-1dry soil,respectively.The highest abundance ratio ofamoAgene between clades B and A was 68.96 in the ZJSX paddy soil and the lowest was 0.62 in the FJNP paddy soil(Fig.1).TheamoAabundance of both clades was consistently strongly affected by soil pH.Regression analysis indicated that theamoAgene abundance of comammoxNitrospiraclades A (R2=0.450 2,P <0.001) and B(R2=0.367 2,P <0.001) was positively related to soil pH (Fig.2).Principal component analysis (PCA) showed that 67.21%of the total variation was explained by principal component(PC)1,while PC 2 explained 21.53%of the total variation,and the two axes together explained 88.74%of the total variation(Fig.3).

Fig.1 Abundance of amoA gene,expressed as log10(No.of amoA gene),of comammox Nitrospira clades A and B and their ratio(clade B/clade A)in 36 paddy soils selected.Vertical bars indicate standard deviations of the means(n=3).

Fig.3 Principal component(PC)analysis of comammox Nitrospira amoA gene abundance and environmental variables in 36 paddy soils selected.MBN=microbial biomass N;MBC=microbial biomass C;TN=total N;TC=total C;SOM=soil organic matter.
Moreover,according to Spearman correlation analysis,the abundance of comammoxNitrospiraclade A was significantly positively correlated(P <0.01)with soil pH,SOM,TC,TN,and latitude and positively correlated(P <0.05)with MBN and longitude(Fig.S1,see Supplementary Material for Fig.S1).The abundance of comammoxNitrospiraclade B was significantly positively correlated(P <0.01)with soil pH,TC,TN,and latitude and positively correlated(P <0.05)with MBN and SOM(Fig.S1).The abundance of comammoxNitrospiraclades A and B was significantly negatively correlated with MAT and MAP (Fig.S1).In addition,there was no significant correlation between comammoxNitrospiraabundance and the other environmental factors(Fig.S1).
α-Diversityof comammox Nitrospira clades A and B
Boxplots were used to exhibit three diversity indices,i.e.,Shannon index,Pielou’s evenness,and Faith’s phylogenetic diversity,of comammoxNitrospiraclades A and B based on ASVs(Fig.4).In this study,the mean Shannon index of comammoxNitrospiraclades A and B was 3.46 and 1.49,respectively,suggesting that clade A had higher diversity than clade B in paddy soils.There were no significant differences in Pielou’s evenness values between the two clades.Furthermore,significant differences in Faith’s phylogenetic diversity of comammoxNitrospiraclades A and B were observed(Fig.4).Spearman correlation analysis between the diversity of comammoxNitrospiraclades A and B and environmental factors showed that comammoxNitrospiraclade B diversity was positively correlated(P <0.05)with TN(Fig.S1).

Fig.4 Boxplots showing the statistical results of α-diversity of comammox Nitrospira clades A and B using three diversity indices,Shannon index(SI),Faith’s phylogenetic diversity(FP),and Pielou’s evenness(PE),in 36 paddy soils selected.***indicates P <0.001.
Community structure of comammox Nitrospira clades A and B
A total of 883 521 and 843 642 high-qualityamoAsequences were obtained from comammoxNitrospiraclades A and B,respectively,in all 36 samples.The number ofamoAsequences from clade A in each sample(24 542 on average)varied from 8 430(HLJHEB)to 84 719(JXYC),and the sequence read length was 375—415 bp,with 98%of the sequence having the length of 415 bp.The number ofamoAsequences from clade B in each sample(23 434 on average)varied from 398(GDJZ)to 40 630(FJSM),and the sequence read length was 375—439 bp;the length of 98%of the sequences was 415 bp(Table SIV,see Supplementary Material for Table SIV).
Redundancy analysis indicated that the community structure of comammoxNitrospiraclades A and B was different among ASVs,and the influencing factors were different(Fig.5).For comammoxNitrospiraclade A,9.73% and 7.46%of the total variation were explained by RDA 1 and RDA 2,respectively(Fig.5a).The community structure of clade A was related(P <0.05)to longitude and SOM.For comammoxNitrospiraclade B,RDA 1 explained 11.65%of the total variation,and RDA 2 explained 6.55% of the total variation (Fig.5b).In comparison,SOM and MBN(P <0.05) were related to the community structure of clade B(Fig.5b).The differences in comammoxNitrospiraclades A and B communities in paddy soils were examined using nonmetric multidimensional scaling analysis based on the Bray-Curtis distance.This demonstrated no significant difference between the comammoxNitrospiraclades A and B communities (Fig.S2,see Supplementary Material for Fig.S2).
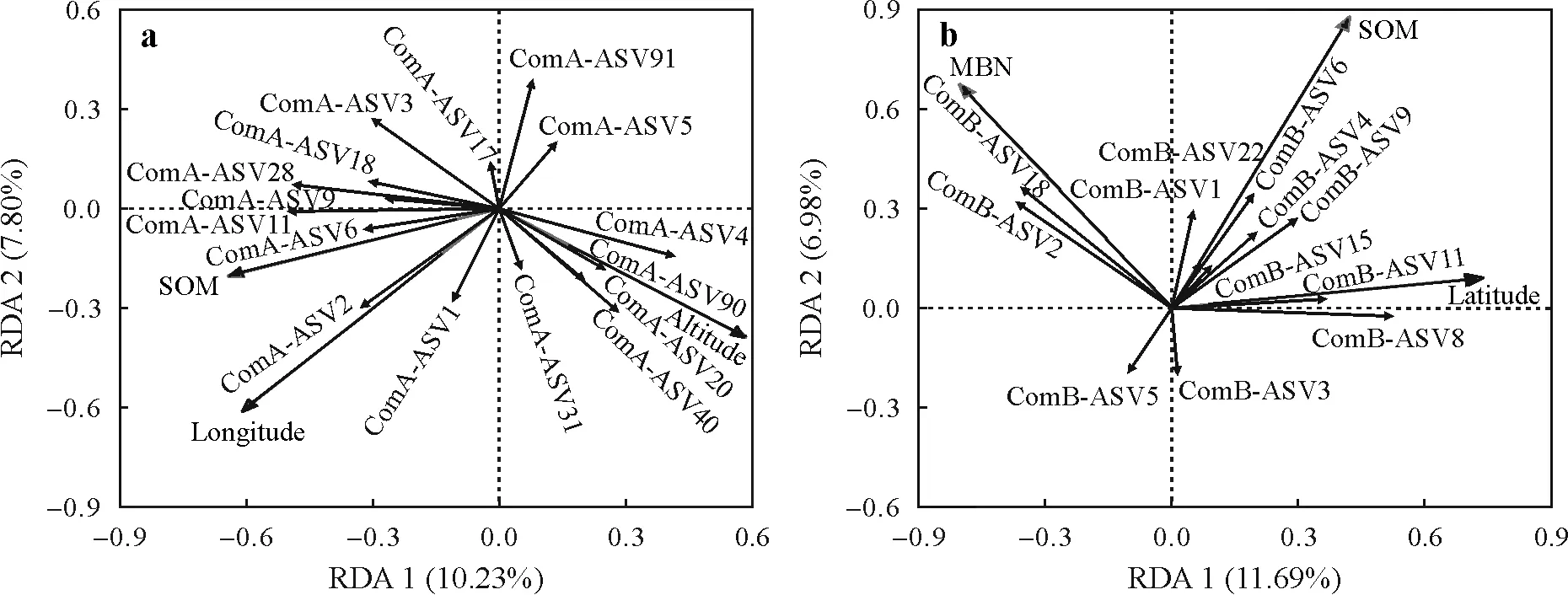
Fig.5 Redundancy analysis(RDA)showing the relationships between the community composition of comammox Nitrospira clades A(a)and B(b)and different environmental factors in 36 paddy soils selected.comA-ASVXXX and comB-ASVXXX are the representative sequences of amoA gene amplicon sequence variants(ASVs)of comammox Nitrospira clades A and B,respectively.SOM=soil organic matter;MBN=microbial biomass N.
To explore the habitat relevance of comammoxNitrospiraclades A and B,the closely relatedamoAgene was searched in the NCBI database.All theamoAgenes belonged toNitrospiraII.Phylogenetic tree based on the existing sequences revealed thatamoAsequences were not only closely related to those from paddy soils and paddy rhizosphere soils but also to the sequences from forests and engineering systems(Fig.6a).The ASVs of comammoxNitrospiraclade A(comA-ASV)were clustered with known sequences from paddy soils,which were significantly different from known sequences from other habitats(Fig.6);this may be attributed to the unique habitats of the paddy fields.In addition,comAASV9 had a close phylogenetic distance withCandidatusNitrospira nitrificans(Fig.6b).Unlike comammoxNitrospiraclade A,the ASVs of comammoxNitrospiraclade B(comBASV)were not clustered with the sequences from a single habitat,but with the sequences from paddy soils,forest soils,and engineering systems (Fig.6).The sequences comBASV6,comB-ASV11,comB-ASV26,and comB-ASV39 were clustered with the sequences from paddy soils and comB-ASV16 and comB-ASV59 with the sequences from acidic forest and neutral paddy soils in China(Fig.6b).In addition,some ASVs were related to biofilters in groundwater wells and circulating aquarium systems(Fig.6b).Surprisingly,comA-ASV96 and comA-ASV5 were dominant in the HLJHEB and SCYB paddy soils,respectively(Fig.7a).Similarly,comB-ASV1,comB-ASV7,comB-ASV13,and comB-ASV39 dominated the comammoxNitrospiraclade B communities in the HLJSH,HLJHEB,SCYB,and GDZJ paddy soils,respectively(Fig.7b).

Fig.6 Neighbor-joining trees for amoA gene amplicon sequence variants(ASVs)of dominant comammox Nitrospira(a)and of dominant comammox Nitrospira clades A and B(representatives with relative abundance of top 1)(b).The sequences in the circles indicate the clustering sequences obtained in this study;those in red and blue circles were from comammox Nitrospira clades A and B,respectively.comA-ASVXXX and comB-ASVXXX are the representative sequences of amoA gene ASVs of comammox Nitrospira clades A and B,respectively.The bootstrap values were 1 000.
Relative influence of environmental factors on abundance,diversityand communitystructure of comammox Nitrospira
Soil pH was the primary factor affecting the abundance of clade A,accounting for approximately 57.41% of the relative influence(Fig.8a).Soil pH,latitude,and MAT were the main factors affecting clade B abundance,accounting for approximately 76.89%of the relative influence(Fig.8b).Soil nitrate,MBC,and MBN mainly affected clade A diversity,accounting for approximately 68.81%of the relative influence(Fig.8c).Soil TC,nitrate,and altitude mainly affected clade B diversity,accounting for approximately 88.37%of the relative influence(Fig.8d).Soil ammonium,SOM,nitrate,and TC were the major factors affecting the clade A community structure,accounting for approximately 46.91%of the relative influence(Fig.8e).Longitude,pH,and MAT explained 8.19%—9.20% of the community structure and thus were not considered main factors influencing the communities of comammoxNitrospiraclade A(Fig.8e).The ABT model also showed that longitude,latitude,TC,and SOM were the most important environmental factors influencing the comammoxNitrospiraclade B community structure,accounting for approximately 46.16%of the relative influence(Fig.8f).Similarly,altitude,pH,and nitrate explained only 8.34%—8.54%of the community structure and were not the major factors affecting the comammoxNitrospiraclade B communities(Fig.8f).The results of Spearman correlation analysis were consistent with the significant correlations observed in the ABT analysis(Fig.S1).

Fig.8 Relative variable influence plots of environmental factors for abundance(a and b),diversity(determined by Shannon index)(c and d),and community structure(determined by Bray-Curtis distance)(e and f)of comammox Nitrospira clades A(a,c,and e)and clade B(b,d,and f)based on aggregated boosted tree models.MBN=microbial biomass N;MBC=microbial biomass C;TN=total N;TC=total C;MAT=mean annual temperature;MAP=mean annual precipitation;SOM=soil organic matter.
DISCUSSION
To our knowledge,this is the first study to combine comammoxNitrospiracommunity structure and abundance with diversity at a large scale in Chinese paddy soils.The qPCR results indicated that both comammoxNitrospiraclades A and B were prevalent in the paddy soils(Fig.1).Previous quantitative studies have shown that in different habitats,AOA,AOB,and comammoxNitrospiracoexist in diverse abundance patterns(Bartelmeet al.,2017;Fowleret al.,2018;Liet al.,2020).Pjevacet al.(2017)have reported that comammoxNitrospirain paddy soils account for 22.9%—48.09% of the total detected autotrophic ammonia-oxidizing microorganisms.In the paddy soils,the Shannon index of comammoxNitrospiraclade A was 2.21—5.39(Fig.4),similar to the values reported for other habitats,including forest soils (2.54),plateau soils (5.33),sediments (2.87—5.03),and water(2.44—4.87)(Xiaet al.,2018).Moreover,the ratio ofamoAgene abundance between clades B and A ranged from 0.62 to 68.96,indicating a much greater gene abundance in clade B than in clade A,except for JSYC and FJNP soils(Fig.1).Other studies have shown that the abundance of comammoxNitrospirais lower than that of AOA and/or AOB in agricultural and forest soils (Pjevacet al.,2017;Kataokaet al.,2018;Liet al.,2020),higher than the abundance of AOA or AOB in acidic forest and grassland soils(Hu and He,2017;Kataokaet al.,2018),or similar to AOA in alkaline forest soils(Hu and He,2017).However,no positive results for comammoxNitrospiraclade B were obtained in terrestrial ecosystems in Australia(Liet al.,2020,2021).In addition,comammoxNitrospiraclade A outnumbered clade B in alkaline paddy soils,with a abundance ratio of clade A/clade B ranging from 6.1 to 43.1(Wanget al.,2018).These results indicate that pH may play an important role in controlling the comammoxNitrospiraabundance.
Although data on comammoxNitrospiraare accumulating rapidly,especially those on comammoxNitrospiraabundance,pH remains to be a significant factor in the control of comammoxNitrospiraabundance in paddy soils.ComammoxNitrospiraclades A and B are highly abundant in forest soils with a gradient of soil pH(Hu and He,2017),and a significant positive correlation was recorded between pH and the abundance of comammoxNitrospira(Liet al.,2020;Luet al.,2020;Xueet al.,2022).The optimal pH for key N-converting enzymes(e.g.,hydroxylamine oxidoreductase and ammonia monooxygenase)is approximately 7.0—8.0(Blumet al.,2018).We found that a variety of factors jointly drive comammoxNitrospira,and pH may be the most important factor affecting the comammoxNitrospiraabundance.Low pH reduces the availability of ammonia,the potential toxic effects of free ammonia(Gubry-Ranginet al.,2011),and the availability of nitrite and increases the toxic effects of nitrous acid(De Boer and Kowalchuk,2001).By sequencing the global soil AOAamoAgene,it was found that the AOA sequences attributable to clusters 14 and 4 were obtained from an environment with pH<5.50 and>7.20,which indicates that the dominant communities of AOA vary with different pH conditions(Da Silvaet al.,2006).The ASVs of comammoxNitrospiraobtained at a certain pH range may correspond to the adaptability of comammoxNitrospirato soil.However,certain environments(e.g.,high temperature,heavy metal contamination,and halophilic or oligotrophic conditions)may contain other undetected comammoxNitrospira.In addition,the adaptability of ammonia-oxidizing microorganisms to soil conditions varies with factors such as pH,water content,substrate concentration,and temperature(Shenet al.,2008;Morimotoet al.,2011;Stevenset al.,2022).
The community structure of clades A and B was affected by both longitude and latitude(Fig.5).Geographical factors have been shown to have a strong influence on AOA and AOB communities(Huet al.,2015;Chenet al.,2017;Liuet al.,2018,2019).Therefore,the relationship between geographic factors(longitude and latitude together explaining 16.40%of clade A variation and 27.53%of clade B variation)and the comammoxNitrospiracommunities was expected(Fig.8e,f).In addition,SOM affected the community diversity of AOA(Chenet al.,2019)and also influenced the comammoxNitrospiracommunity (explaining 11.37% of clade A variation and 9.02% of clade B variation) (Fig.8e,f).Under different ammonium concentrations,both AOA and AOB exhibited clear community changes(Huet al.,2012).Ammonium showed a strong relationship with the comammoxNitrospiraclade A community (explaining 15.00%variation),which was higher than that with comammoxNitrospiraclade B community(explaining 7.20%variation)(Fig.8e,f).These relationships between environmental factors and comammoxNitrospiracommunity may be due to the fact that comammoxNitrospiraclades A and B contain two different types of ammonia transporters(Palomoet al.,2018),resulting in different ammonia absorption characteristics.The Rh-type ammonia transporter (e.g.,AOB and comammoxNitrospiraclade A)has ammonia affinity and absorption in the millimolar range,whereas the MEP-type ammonia transfer protein(e.g.,AOA and comammoxNitrospiraclade B)has a higher ammonia affinity and absorption capacity in the micromolar range(Palomoet al.,2018).For comammoxNitrospiraclade A,ammonium was the primary influencing factor of the community.This may be due to the long-term drying-wetting alternation of the rice field,which causes ammonium to occur at the millimolar level.At this level,comammoxNitrospiraclade A had a higher ammonium affinity and absorption capacity.
Ammonium was the primary influencing factor of the comammoxNitrospiraclade A community,but its effect on comammoxNitrospiraclade B was weaker.Nonmetric multidimensional scaling analysis showed that there was no clear separation between the comammoxNitrospiraclade A communities and no obvious separation between the comammoxNitrospiraclade B communities(Fig.S2).Although the higher affinity of comammoxNitrospirafor substrate ammonia may enhance their ability to compete with other microorganisms in soil with an extremely low ammonia concentration,this does not mean that the comammoxNitrospiracommunity has obvious environmental separation.As the combined effect of soil pH,temperature,and fertilization also affects the ammonia oxidation process,there is no significant difference in the community composition of comammox(Wanget al.,2017;Liet al.,2018).Soil pH is closely related to the overall ammonia oxidizer community(Huet al.,2013,2015),and the large-scale biogeographic pattern is mainly controlled by soil pH,geographic distance,and climatic factors(Huet al.,2015).Comparative genomic analysis has shown that there are significant differences between comammoxNitrospiraclades A and B (Palomoet al.,2018).These differences indicate that environmental factors such as soil pH,temperature,and TN may affect the functional importance of comammoxNitrospirain natural habitats(Aiet al.,2013).This may be because the factors that adjust microbial communities are usually intricate and involve multiple environmental variables(Huet al.,2015).Therefore,to better understand the molecular mechanism and relative influence of comammoxNitrospira,it is necessary to conduct more research,especially on pure cultures and metagenomics.
CONCLUSIONS
Our results provide new evidence that comammoxNitrospiraare widely distributed in Chinese paddy soils,suggesting their potential functions in soil nitrification,which can be influenced by a range of factors.The abundance of comammoxNitrospiraclade A was mostly lower than that of clade B,whereas its diversity was mostly higher than that of clade B.Phylogenetic analysis showed that the ASVs were clustered with three types of comammoxNitrospirathat are currently purely cultured.There was a clear relationship between the comammoxNitrospiracommunity and the habitat conditions.Some ASVs had a unique dominant position in certain habitats.These findings can help to clarify the process of ammonia oxidation by ammonia-oxidizing microorganisms and explain how their distinct preferences for environmental factors and habitat define the relative contributions of comammoxNitrospiraclades A and B to soil nitrification.
ACKNOWLEDGEMENT
This work was supported by the National Natural Science Foundation of China(Nos.42077036 and 41877051)and the Ningbo Municipal Science and Technology Bureau,China(No.202002N3079).
SUPPLEMENTARY MATERIAL
Supplementary material for this article can be found in the online version.
- Pedosphere的其它文章
- Carbon farming by recarbonization of agroecosystems
- Root exclusion methods for partitioning of soil respiration:Review and methodological considerations
- Utilization of lignocellulosic plant residues for compost formation and its role in improving soil fertility
- A critical review of microbially induced carbonate precipitation for soil stabilization:The global experiences and future prospective
- Hand-feel soil texture observations to evaluate the accuracy of digital soil maps for local prediction of soil particle size distribution:A casestudy in Central France
- Influence of soil physicochemical properties,particle size fractions and mineralogy on the leaching potentials of arsenic and antimony in abandoned mine soils

