前列腺癌的DNA甲基化及其临床应用
赵帆, 杨泽
北京大学医学部第五临床医学院, 卫生部北京医院老年医学研究所, 北京 100730
前列腺癌的DNA甲基化及其临床应用
赵帆, 杨泽
北京大学医学部第五临床医学院, 卫生部北京医院老年医学研究所, 北京 100730
目前认为恶性肿瘤的形成是遗传和表观遗传机制共同作用的结果。表观遗传机制包括DNA甲基化、组蛋白修饰和 miRNA。DNA异常甲基化(高甲基化和低甲基化)是前列腺癌最具特征的表观遗传改变, 它能够导致基因组不稳定, 调控基因的异常表达, 在前列腺癌的形成和发展中起到重要作用。同时, DNA甲基化作为前列腺癌表观遗传研究的一个热点, 为临床前列腺癌的早期诊断、预后评估及药物治疗提供新的方法和途径。文章根据前列腺癌的 DNA高甲基化和低甲基化的最新研究成果阐述了前列腺癌形成的表观遗传学机制, 并且讨论了它们在前列腺癌临床转化方面的最新研究进展。
前列腺癌; DNA 高甲基化; DNA 低甲基化
前列腺癌是老年男性最常见的恶性肿瘤之一。目前, 前列腺癌已成为中老年男性癌症发病的第 2大病因, 位居男性癌症致死人数的第6位[1]。前列腺癌的发病机制较为复杂, 目前认为遗传和表观遗传机制共同作用导致前列腺癌的发生、发展, 其中表观遗传在前列腺癌的形成中起到重要的作用。表观遗传是指在染色体 DNA序列不发生改变的情况下产生的一种可稳定遗传的表型[2]。表观遗传机制包括DNA甲基化、组蛋白修饰和miRNA, 它们分别通过转录前和转录后控制基因表达, 其中DNA甲基化在前列腺癌表观遗传机制研究中成果最多, 也最为引人注目。在哺乳动物基因组中, DNA甲基化通常发生在 CpG双核苷酸的胞嘧啶上, 由硫-腺苷-甲硫氨酸(S-adenosylmethionine, SAM)提供甲基供体,在DNA甲基转移酶(DNA mthyltransferase, DNMT)的催化下, 将甲基转移到CpG双核苷酸胞嘧啶的第5个碳原子上。CpG不是随机分布的, 它最常见于基因组CpG岛的位置, 哺乳动物中一半以上的基因都含有CpG岛, 大部分CpG岛位于基因启动子、非编码区和第一外显子, 且在正常细胞内不发生甲基化[3,4]。前列腺癌中 DNA异常甲基化主要表现为基因组广泛低甲基化和局部基因启动子区域的高甲基化。DNA异常甲基化发生在前列腺癌的形成过程中, 且DNA甲基化能够通过药物发生逆转, 因此, 前列腺癌 DNA甲基化的早期筛查及前列腺癌去甲基化药物的临床应用, 可能会为临床早期诊断和治疗前列腺癌提供新的思路。
本文主要阐述了前列腺癌表观遗传机制中 DNA异常甲基化的最新研究成果以及前列腺癌 DNA异常甲基化在临床转化中的应用及存在的问题。
1 DNA高甲基化
基因组中 DNA高甲基化常发生于基因的启动子区域, 即富含CpG的CpG岛区域。这些区域在正常细胞中通常是非甲基化的。这些基因主要参与激素应答, 细胞增殖、迁移和侵袭, DNA修复及转录调控等(表1)。基因启动子DNA高甲基化致使相关基因表达沉默是前列腺肿瘤形成的一个重要原因。根据它们的功能和信号通路不同, 主要包括以下相关基因:
1.1 激素应答相关基因
雄激素受体(Androgen receptor, AR)是类固醇激素受体家族的一个成员, 与雄激素结合后与辅助蛋白分离进入细胞核内, 刺激雄激素应答基因的转录。5-氮脱氧胞苷(5-aza-CdR)可逆转前列腺癌干细胞由AR基因启动子DNA高甲基化导致的表达沉默, AR表达上调可降低前列腺癌干细胞特性, 诱导癌细胞的增殖和分化[5]。视黄酸受体 β(Retinoic acid receptor beta, RARB)是甲状腺类固醇激素受体家族成员之一, 它与具有生物活性的维生素 A-视黄酸结合, 参与细胞生长和分化及胚胎形成过程中的信号转导。RARB基因启动子区域DNA高甲基化可发生在多个肿瘤的形成过程中, 如前列腺癌[6,7]、乳腺癌[8]、肺癌[9]、食管癌[10]、甲状腺癌[11]、膀胱癌[12]、结直肠癌[13]、恶性胶质瘤[14]、鼻咽癌[15]等。因此, 我们推断该基因可能在多个肿瘤形成过程中参与调节肿瘤形成的共同传导途径。G蛋白偶联受体(G protein coupling receptors, GPCRs)能够刺激AR的雄激素非依赖性激活, 是导致激素难治性前列腺癌的发生的重要因素。G 蛋白信号调节因子 2 (Regulator of G-protein signaling 2, RGS2)是一种GTP酶激活蛋白,能够抑制 GPCRs, 介导骨髓细胞分化, 可能参与白血病的形成。RGS2基因启动子DNA高甲基化异常能够导致雄激素非依赖性前列腺癌细胞生长, 表明RGS2基因可能通过调控GPCRs参与AR反式激活通路[16]。ATP结合盒亚家族成员 1(ATP-binding cassette, sub-family A, member 1, ABCA1)是存在于细胞膜表面的外流性转运蛋白, 能够转运细胞内多余的胆固醇, 在维持细胞胆固醇稳态方面起到重要作用。ABCA1基因启动子DNA高甲基化导致基因表达沉默, 它能使前列腺细胞内的胆固醇升高, 雄激素合成增加, 后者通过AKT信号通路促进前列腺癌的恶性进展[17]。
1.2 抑癌基因
在前列腺癌DNA高甲基化研究中, 最常见的是抑癌基因启动子 DNA高甲基化。癌甲基化蛋白 1 (Hypermethylated in cancer 1, HIC1)基因表达一种转录阻抑蛋白, 在细胞中发挥生长调控和抑癌基因的作用。前列腺癌细胞系、前列腺组织和血浆中均发现HIC1基因启动子DNA高甲基化, 在前列腺癌细胞异种移植的小鼠体内诱导表达沉默的 HIC1基因激活, 可以观察到它具有抑制前列腺肿瘤生长、迁移和侵袭的作用[18,19]。结肠腺瘤性息肉 (Adeno-matous polyposis coli, APC) 基因表达一种WNT信号通路拮抗剂, 它参与细胞的迁移、侵袭、转录激活和细胞凋亡, 是一种常见的抑癌基因, 该基因突变常导致家族性结肠腺瘤性息肉病。APC基因启动子区域 DNA高甲基化在前列腺患者组织中常见[20],且甲基化程度与前列腺癌肿瘤分期和Glison评分呈正相关[21]。WNT抑制因子1(WNT inhibitory factor 1, WIF1) 基因编码一种胞外信号分子, 能够抑制WNT蛋白, 参与胚胎发育。该基因启动子DNA高甲基化发生在大多数前列腺癌细胞系中, 体外诱导PC-3细胞系表达 WIF1, 可降低细胞迁移和侵袭能力, 上调E-钙粘素(E-cadherin, CDH1)、角蛋白-8,18 (Keratin-8 and-18, KRT8,18)的表达, 从而抑制上皮细胞向间充质细胞转化。在异种移植小鼠模型发现WIF1表达升高能够抑制前列腺肿瘤生长[22]。原钙粘附蛋白10(Protocadherin 10, PCDH10)基因属于原钙黏蛋白家族成员, 为抑癌基因, 编码钙粘素相关蛋白受体, 参与脑内特定细胞粘附及其功能联系, 也参与前列腺癌的发生、发展[23]。
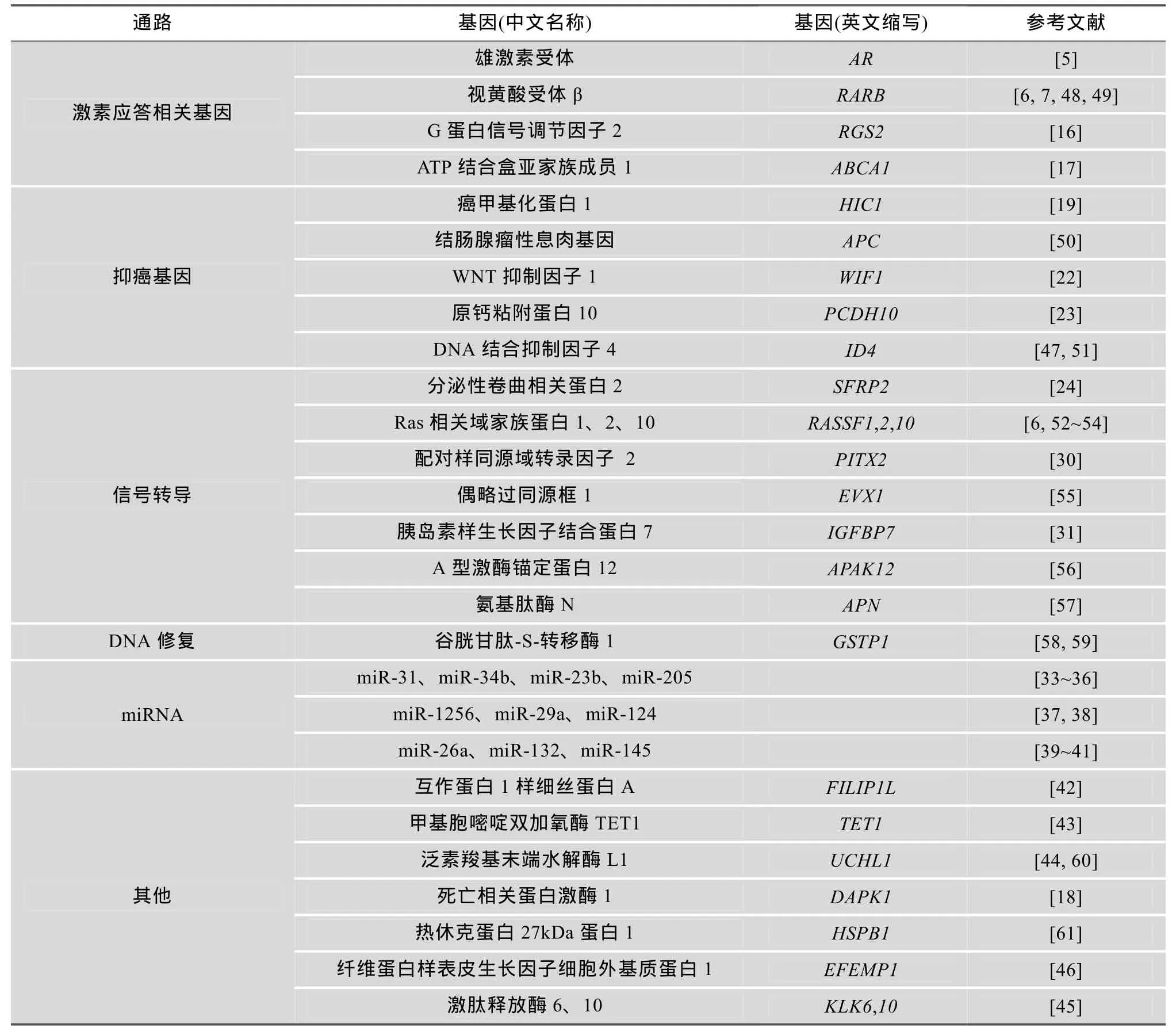
表1 前列腺癌中启动子区域发生DNA高甲基化的基因
1.3 信号转导基因
WNT信号通路过度激活与肿瘤发生和肿瘤侵袭相关, 分泌性卷曲相关蛋白 2(Secreted frizzledrelated protein 2, SFRP2)基因在WNT信号通路中能够抑制该信号通路过度激活。SFRP2基因启动子DNA高甲基化在前列腺癌组织中的发生率明显高于癌旁、高分级前列腺上皮内瘤和前列腺增生组织[18,24]。Ras相关域家族蛋白 1(Ras association domain family member1, RASSF1)基因编码一种与Ras效应蛋白相似的蛋白, 该基因启动子DNA 高甲基化可在多个肿瘤组织中检测到, 如前列腺癌[21]、乳腺癌[25]、膀胱癌[26]、肝癌[27]、非小细胞肺癌[28]、卵巢癌[29]等, 该基因同 RARB基因一样, 在肿瘤形成过程中参与其共同通路的调节。配对样同源域转录因子 2(Paired-like homeodomain 2, PITX2)基因表达一种转录因子, 调控原骨胶原赖氨酸羟化酶(Procollagenlysyl hydroxylase)基因的表达, 在促生长激素细胞和催乳素细胞的末端分化中发挥作用,同时也参与眼、牙齿和腹部器官的发育。在前列腺癌细胞系P69和M12中PITX2基因启动子区域均被甲基化, 它可能作为AR和IGF-1R基因上游的调节因子, 通过异常调节AR和IGF1-R通路, 影响前列腺细胞的正常生长[30]。胰岛素样生长因子蛋白7(Insulin-like growth factor binding protein 7, IGFBP7)能够与胰岛素生长因子(Insulin-like growth factor, IGF)结合, 参与前列环素的合成及细胞粘附。IGFBP7基因启动子DNA高甲基化在多种前列腺癌细胞系和组织中检测到[31], 但目前其作用机制尚不清楚。
1.4 DNA修复基因
谷胱甘肽 S-转移酶 1(Glutathione S-transferase pi 1, GSTP1)基因, 属于谷胱甘肽S-转移酶基因家族成员, 它通过催化疏水性和亲电性基团与还原型谷胱甘肽结合发挥细胞解毒作用。在对25例行前列腺切除术的前列腺癌、癌旁基因甲基化水平评估后,发现GSTP1基因的甲基化水平在癌组织中明显高于癌旁组织[20]。研究发现:前列腺癌细胞内GSTP1基因启动子 DNA甲基化所致的表达沉默使胞内活性氧物质(ROS)聚集, DNA损伤标记物——胞内羟基脱氧鸟苷(8-oxo-2′-deoxogunosine, 8-OHdG)增加, GSTP1基因表达缺失可增加正常前列腺细胞对氧化应激诱导的DNA损伤的敏感性, 从而导致前列腺癌形成[32]。
1.5 miRNA
miRNA是内源性非编码的 RNA, 能够与靶mRNA3′-UTR(Untranslated region)部分互补结合抑制其翻译或诱导特定的靶mRNA降解。前列腺癌中部分miRNA的异常调控也是因为表达miRNA的基因启动子区域发生DNA高甲基化。在前列腺癌中启动子区域DNA高甲基化导致miR-31表达沉默, AR表达升高, 可能是前列腺癌的恶性进展病因学机制之一[33]。miR-34b和miR-23b都具有抑制细胞增殖、迁移和侵袭, 以及 EMT(上皮间质转化)的作用, miR-34b和 miR-23b基因高甲基化导致其表达降低,原癌基因 Scr激酶表达升高, 前列腺肿瘤细胞恶性增殖, 导致患者复发生存期缩短[34,35]。此外, miR-205、miR-29a、miR-1256、miR-124、miR-26a、miR-132、miR-145基因高甲基化也参与前列腺癌形成[36~41]。
1.6 其他
互作蛋白1样细丝蛋白A(Filamin A interacting protein 1-like, FILIP1L)基因表达一种细胞血管内皮活性调控因子, FILIP1L基因启动子高甲基化在前列腺癌中常见, 可能与前列腺癌形成过程中肿瘤血管的形成相关[42]。甲基胞嘧啶双加氧酶 TET1(Tetmethylcytosinedioxygenase 1, TET1)基因表达参与胞嘧啶脱甲基的脱甲基酶。TET1基因启动子DNA高甲基化, 其 mRNA表达降低, 能够下调金属蛋白酶抑制剂1、2(Tissue inhibitor of metalloproteinase 1 and 2, TIMP1、TIMP2)表达, 从而促进前列腺癌转移、侵袭[43]。死亡相关蛋白激酶1(Death-associated protein kinase1, DAPK1)基因参与γ干扰素(INF-γ)诱导的程序性细胞凋亡。DAPK1基因启动子区域DNA甲基化在前列腺癌组织中的发生率明显高于正常前列腺组织[18]。泛素羧基末端水解酶L1(Ubiquitin carboxyl-terminal esterase L1, UCHL1)基因参与泛素化过程, 能够水解泛素羧基末端的甘氨酸, 同时参与细胞增殖和分化。UCHL1基因启动子DNA甲基化在前列腺癌组织中的发生率为90%, 癌旁组织为15%,差异明显[44]。此外, 激肽释放酶 6、10(Kallikreinrelated peptidase 6,10, KLK6,10)、DNA结合蛋白抑制因子4(Inhibitor of DNA binding 4, ID4)、锌指蛋白132(Zinc finger protein 132, ZNF132)、A型激酶铆钉蛋白12(A-kinase anchor protein 12, AKAP12)、纤维蛋白样表皮生长因子细胞外基质蛋白 1(EGF containing fibulin-like extracellular matrix protein 1, EFEMP1)等基因在前列腺癌细胞中也发生 DNA高甲基化, 可能通过其他途径参与前列腺癌的发生、发展[45~47], 具体机制目前尚不清楚。
2 DNA低甲基化
尽管前列腺癌基因组中的部分基因启动子区域DNA常发生CpG高甲基化, 但是在前列腺癌基因组中, CpG 位点低甲基化却占明显优势[62], 在前列腺癌基因组内呈现广泛的低甲基化, 而且DNA低甲基化并非随机的, 这些CpG位点在正常前列腺组织中为低甲基化, 它们常位于印记基因、逆转录转座子、内源性病毒序列、基因组的内含子、基因间区以及基因组中散在分布的重复序列或端粒的重复序列内[63]。基因组中该位置的CpG位点低甲基化会导致印记基因过表达、染色质结构改变、表观遗传重组及基因组不稳定, 在前列腺癌的发病机制中同样发挥重要作用。目前对前列腺癌DNA低甲基化的研究相对较少, 可能由于DNA低甲基化通常发生在重复元件内, 且DNA序列会有部分重叠, 因此较难在实验中研究。
长散在核重复序列(Long interspersed nuclear element1, LINE1)为基因组内散在分布的重复序列,在正常细胞基因组内通常是高甲基化的。在前列腺癌样本中发生低甲基化, 且更常见于转移性前列腺癌[64]。此外, 胰岛素样生长因子(Insulin-like growth factor 2, IGF2)基因为印记基因, 其父源等位基因表达相应蛋白, 参与机体的生长和发育。IGF2基因低甲基化导致两个等位基因同时表达, 前列腺癌中IGF2基因印记控制区尤其是CTCF结合域的甲基化水平与前列腺增生组织有显著性差异[65,66]。三叶因子(Trefoil factor, TFF)基因在胃黏膜中表达一种稳定的分泌性蛋白。前列腺癌细胞系中TFF1、3基因甲基化水平明显低于正常前列腺细胞, 但它们在前列腺肿瘤的形成中的作用尚不清楚[67]。
3 DNA甲基化的前列腺癌早期诊断
表观遗传标记物, 尤其是DNA甲基化, 可能会成为未来临床检测和诊断前列腺癌的新的方法和手段, 主要因为表观遗传改变在前列腺癌中普遍存在,而且在前列腺肿瘤形成前期就已经发生, 有利于临床早期筛查和检测。其次, 相较于 RNA检测来说,基因组DNA的检测相对稳定, 而且检测方法多样。再次, 随着DNA甲基化检测技术的不断发展, 标准化的高通量检测平台的建立能够用于多个样本DNA 甲基化多个位点的检测, 有利于临床开展DNA甲基化检测。另外, DNA甲基化作为标志物检测手段多样, 不仅在肿瘤组织中检测, 还可以在体液(如尿液、血液)中检测。
目前, 很多研究致力于相关基因启动子DNA高甲基化作为前列腺癌生物标志物的临床检测。定量焦磷酸测序的方法对52例前列腺增生组织和97例前列腺癌组织中APC基因和GSTP1基因两种DNA甲基化水平进行分析, 发现区分前列腺癌和前列腺增生的敏感性为92.8%, 特异性为100%[68], 在对30多个前列腺癌GSTP1基因启动子DNA高甲基化研究进行Meta分析, 发现GSTP1基因启动子DNA甲基化检测能够提高临床前列腺癌诊断特异性[69]。APC基因 DNA甲基化检测可以作为首次活检阴性的前列腺癌高危人群再次活检的指标[70]。对34例早期前列腺癌的患者的尿沉渣进行 DNA甲基化分析发现, RARB和RASSF1基因启动子DNA高甲基化的检出率分别为71%和44%[6]。此外, 联合检测EVX1和成纤维细胞生长因子1(Fibroblast growth factor 1, FGF1)基因启动子DNA高甲基化可进一步鉴别出前列腺穿刺活检结果为阴性的前列腺癌患者, 减少检查者不必要的痛苦, 也能够降低检测成本和穿刺后并发症[71]。
基因 DNA甲基化检测作为临床前列腺癌的早期诊断, 可以提高前列腺癌诊断特异性, 为首次活检阴性的前列腺癌患者是否二次活检提供临床诊断参考。但前列腺癌DNA甲基化检测作为新的诊断方法也有需要待解决的问题:一是如何选择前列腺癌特异性的DNA异常甲基化基因, 有利于提高诊断特异性; 二是如何提高前列腺癌特异性的DNA异常甲基化基因检测的敏感性。
4 DNA甲基化对前列腺癌治疗
与常见的遗传改变不同, 表观遗传学的改变不涉及到DNA序列中碱基的改变。这种表观遗传学的可行性使它们有望成为潜在的药物治疗靶。目前主要的DNMT抑制剂分为两大类:核苷类似物和非核苷类似物。它们通过抑制DNMT来恢复相关基因的表达功能, 从而达到治疗前列腺癌的目的。前者主要有 5-氮杂胞苷、5-氮脱氧胞苷和 zebularine(Zeb,化学名 1-(β-D-呋喃核糖苷)-1,2-二氢嘧啶-2-酮), 前两种药物已经被美国FDA批准用于治疗骨髓异常增生综合征(MDS)。Zebularine能够使LNCaP和DU145前列腺癌细胞系GSTP1基因发生去甲基化, 提高其他化疗药物的抗肿瘤活性[72]。
后者主要包括从植物中提取化学物质如大豆异黄酮、姜黄素、茶多酚[73]、mahanine[74]、kazinol Q[75]、disulfiram[76]等, 最新合成的甲基化抑制剂RG108[77]及其他化学物质如反式维甲酸等。大豆异黄酮能够抑制前列腺癌细胞系中的 DNMT, 使多个基因启动子DNA发生去甲基化, 抑制前列腺癌细胞生长和侵袭的作用[37,78,79]。在TRAMP小鼠前列腺癌表观遗传学的研究中发现, 姜黄素可以使启动子高甲基化的Nrf2基因发生去甲基化[80]。全反式维甲酸能够使R阴性的前列腺细胞系 DU145中表达沉默的HOXB13基因的甲基化水平降低, 从而抑制细胞增殖[81]。
上述DNMT抑制剂在前列腺癌甲基化的研究中主要应用于细胞系, 之所以没有应用于临床, 一是目前体外研究结果还未对DNMT抑制剂在抑制前列腺癌 DNA高甲基化方面提供一个可靠且公认的生物学机制; 二是目前还没有一种DNMT抑制剂为靶向治疗, DNMT抑制剂的应用可能会干扰正常组织及细胞功能, 这种长期作用的结果是未知的; 三是在临床前期实验中, 这些药物对实体肿瘤细胞的细胞毒性较大及反应率较低, 存在的药物副作用远远大于药物的治疗作用, 因此限制了DNMT抑制剂在前列腺癌临床治疗中的应用。
5 DNA甲基化对前列腺癌的预后评估
DNA甲基化检测对前列腺癌经临床治疗后疾病的预后预测也同样重要。对267例根治性前列腺癌切除术患者和 111例前列腺癌保守治疗患者组织APN基因启动子区域甲基化分析、免疫组化分析及随访研究发现, APN基因启动子区域高甲基化明显缩短前列腺癌患者的复发生存期和肿瘤生存期[57]。经根治性前列腺癌切除术患者通过使用 RT-PCR方法检测PIX2基因高甲基化, 能够预测前列腺癌患者PSA 复发, 是一个潜在的前列腺癌预后标志物[82]。低Gleason评分患者中HSPB1基因甲基化可以作为不良预后的生物标志物[61]。此外, 术前PSA水平低的前列腺癌患者 miR-205基因启动子高甲基化可能与PSA复发相关[36]。EVX1基因在前列腺癌中能够预测PSA复发[55]。
6 结语与展望
表观遗传改变, 尤其是基因启动子区域的DNA高甲基化是前列腺癌的常见特征, 在前列腺癌的发生和发病机制的进展中起到重要作用。但是, 前列腺癌 DNA异常甲基化研究还存在很多问题。第一,大部分研究只是针对前列腺癌中某个特定基因启动子区域中的一个或几个 CpG位点进行甲基化分析,整个基因启动子区域的甲基化水平研究不够充分;第二, 现存的技术手段很难对前列腺癌DNA异常甲基化进行精确的定量研究; 第三, 需要更多的前列腺癌组织样本进行大样本的重复验证; 第四, 表观遗传中DNA甲基化、组蛋白修饰及miRNA三者在癌症的发生、发展中是相互作用的, 因此需要更多更广泛的研究全面的阐述前列腺癌表观遗传机制;此外, 遗传和表观遗传共同作用导致前列腺癌的发生, 因此, 如何将遗传和表观遗传结合起来共同解释前列腺癌发病机制也是前列腺癌研究需解决的一个问题。
随着科学技术及研究水平的不断提高, 我们相信通过获得大量的前列腺肿瘤特异性的遗传和表观遗传学改变, 尤其是前列腺癌DNA甲基化的改变, 从而对前列腺癌的发病机制进行更全面的阐述, 并以此作为生物学标志物, 可能会为前列腺癌的临床早期检测、诊断、预后评估及随访提供新的方法和手段。
[1] Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, De Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R,Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, Macintyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, Mcanulty JH, Mcdermott MM, Mcgrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O'donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA, 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, Almazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet, 2012, 380(9859): 2095–2128.
[2] Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev, 2009, 23(7): 781–783.
[3] Bird AP. CpG-rich islands and the function of DNA methylation. Nature, 1986, 321(6067): 209–213.
[4] Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet, 1989, 83(2): 181–188.
[5] Tian J, Lee SO, Liang L, Luo J, Huang CK, Li L, Niu YJ, Chang C. Targeting the unique methylation pattern of androgen receptor (AR) promoter in prostate stem/progenitor cells with 5-aza-2′-deoxycytidine (5-AZA) leads to suppressed prostate tumorigenesis. J Biol Chem, 2012, 287(47): 39954–39966.
[6] Daniūnaite K, Berezniakovas A, Jankevičius F, Laurinavičius A, Lazutka JR, Jarmalaite S. Frequent methylation of RASSF1 and RARB in urine sediments from patients with early stage prostate cancer. Medicina (Kaunas), 2011, 47(3): 147–153.
[7] Tang DL, Kryvenko ON, Mitrache N, Do KC, Jankowski M, Chitale DA, Trudeau S, Rundle A, Belinsky SA, Rybicki BA. Methylation of the RARB gene increases prostate cancer risk in black Americans. J Urol, 2013, 190(1): 317–324.
[8] Mirza S, Sharma G, Parshad R, Srivastava A, Gupta SD, Ralhan R. Clinical significance of promoter hypermethylation of ERbeta and RARbeta2 in tumor and serum DNA in Indian breast cancer patients. Ann Surg Oncol, 2012, 19(9): 3107–3115.
[9] Feng QH, Hawes SE, Stern JE, Wiens L, Lu H, Dong ZM, Jordan CD, Kiviat NB, Vesselle H. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev, 2008, 17(3): 645–654.
[10] 王长春, 毛伟敏, 凌志强. 维甲酸受体 β2和 p16INK4α基因甲基化在食管鳞癌患者肿瘤组织和外周血中的表达. 中华肿瘤杂志, 2012, 34(6): 441–445.
[11] Brait M, Loyo M, Rosenbaum E, Ostrow KL, Markova A, Papagerakis S, Zahurak M, Goodman SM, Zeiger M, Sidransky D, Umbricht CB, Hoque MO. Correlation between BRAF mutation and promoter methylation of TIMP3, RARbeta2 and RASSF1A in thyroid cancer. Epigenetics, 2012, 7(7): 710–719.
[12] Chan MW, Chan LW, Tang NL, Tong JH, Lo KW, Lee TL, Cheung HY, Wong WS, Chan PS, Lai FM, To KF. Hypermethylation of multiple genes in tumor tissues and voided urine in urinary bladder cancer patients. Clin Cancer Res, 2002, 8(2): 464–470.
[13] Miladi-Abdennadher I, Abdelmaksoud-Damak R, Ayadi L, Khabir A, Frikha F, Kallel L, Amouri A, Frikha M, Sellami-Boudawara T, Gargouri A, Mokdad-Gargouri R. Hypermethylation of RARbeta2 correlates with high COX-2 expression and poor prognosis in patients with colorectal carcinoma. Tumour Biol, 2010, 31(5): 503–511.
[14] Piperi C, Themistocleous MS, Papavassiliou GA, Farmaki E, Levidou G, Korkolopoulou P, Adamopoulos C, Papavassiliou AG. High incidence of MGMT and RARbeta promoter methylation in primary glioblastomas: association with histopathological characteristics, inflammatory mediators and clinical outcome. Mol Med, 2010, 16(1–2): 1–9.
[15] Fendri A, Masmoudi A, Khabir A, Sellami-Boudawara T, Daoud J, Frikha M, Ghorbel A, Gargouri A, Mokdad-Gargouri R. Inactivation of RASSF1A, RARbeta2 and DAP-kinase by promoter methylation correlates withlymph node metastasis in nasopharyngeal carcinoma. Cancer Biol Ther, 2009, 8(5): 444–451.
[16] Wolff DW, Xie Y, Deng CS, Gatalica Z, Yang MJ, Wang B, Wang JC, Lin MF, Abel PW, Tu YP. Epigenetic repression of regulator of G-protein signaling 2 promotes androgenindependent prostate cancer cell growth. Int J Cancer, 2012, 130(7): 1521–1531.
[17] Lee BH, Taylor MG, Robinet P, Smith JD, Schweitzer J, Sehayek E, Falzarano SM, Magi-Galluzzi C, Klein EA, Ting AH. Dysregulation of cholesterol homeostasis in human prostate cancer through loss of ABCA1. Cancer Res, 2013, 73(3): 1211–1218.
[18] Kilinc D, Ozdemir O, Ozdemir S, Korgali E, Koksal B, Uslu A, Gultekin YE. Alterations in promoter methylation status of tumor suppressor HIC1, SFRP2, and DAPK1 genes in prostate carcinomas. DNA Cell Biol, 2012, 31(5): 826–832.
[19] Zheng JH, Wang JL, Sun XQ, Hao MG, Ding T, Xiong D, Wang XM, Zhu Y, Xiao G, Cheng GC, Zhao MZ, Zhang J, Wang JH. HIC1 modulates prostate cancer progression by epigenetic modification. Clin Cancer Res, 2013, 19(6): 1400–1410.
[20] Steiner I, Jung K, Schatz P, Horns T, Wittschieber D, Lein M, Dietel M, Erbersdobler A. Gene promoter methylation and its potential relevance in early prostate cancer diagnosis. Pathobiology, 2010, 77(5): 260–266.
[21] Liu LY, Kron KJ, Pethe VV, Demetrashvili N, Nesbitt ME, Trachtenberg J, Ozcelik H, Fleshner NE, Briollais L, Van Der Kwast TH, Bapat B. Association of tissue promoter methylation levels of APC, TGFbeta2, HOXD3 and RASSF1A with prostate cancer progression. Int J Cancer, 2011, 129(10): 2454–2462.
[22] Yee DS, Tang YX, Li XS, Liu ZB, Guo Y, Ghaffar S, Mcqueen P, Atreya D, Xie J, Simoneau AR, Hoang BH, Zi XL. The Wnt inhibitory factor 1 restoration in prostate cancer cells was associated with reduced tumor growth, decreased capacity of cell migration and invasion and a reversal of epithelial to mesenchymal transition. Mol Cancer, 2010, 9: 162.
[23] Li ZS, Li WJ, Xie J, Wang Y, Tang AF, Li XX, Ye JX, Gui YT, Cai ZM. Epigenetic inactivation of PCDH10 in human prostate cancer cell lines. Cell Biol Int, 2011, 35(7): 671–676.
[24] Perry AS, O'hurley G, Raheem OA, Brennan K, Wong S, O'grady A, Kennedy AM, Marignol L, Murphy TM, Sullivan L, Barrett C, Loftus B, Thornhill J, Hewitt SM, Lawler M, Kay E, Lynch T, Hollywood D. Gene expression and epigenetic discovery screen reveal methylation of SFRP2 in prostate cancer. Int J Cancer, 2013, 132(8): 1771–1780.
[25] Xu J, Shetty PB, Feng WW, Chenault C, Bast RC Jr, Issa JP, Hilsenbeck SG, Yu YH. Methylation of HIN-1, RASSF1A, RIL and CDH13 in breast cancer is associated with clinical characteristics, but only RASSF1A methylation is associated with outcome. BMC Cancer, 2012, 12: 243.
[26] Gao TY, Wang SK, He BS, Pan YQ, Song GQ, Gu L, Chen LP, Nie ZL, Xu YQ, Li R. The association of RAS association domain family Protein1A (RASSF1A) methylation states and bladder cancer risk: a systematic review and meta-analysis. PLoS ONE, 2012, 7(11): e48300.
[27] Feng Y, Xue WJ, Li P, Sha ZY, Huang H, Rui L, Li HX, Mao QS. RASSF1A hypermethylation is associated with aflatoxin B1 and polycyclic aromatic hydrocarbon exposure in hepatocellular carcinoma. Hepatogastroenterology, 2012, 59(118): 1883–1888.
[28] Wang J, Wang BC, Chen X, Bi JW. The prognostic value of RASSF1A promoter hypermethylation in non-small cell lung carcinoma: a systematic review and meta-analysis. Carcinogenesis, 2011, 32(3): 411–416.
[29] Bondurant AE, Huang ZQ, Whitaker RS, Simel LR, Berchuck A, Murphy SK. Quantitative detection of RASSF1A DNA promoter methylation in tumors and serum of patients with serous epithelial ovarian cancer. Gynecol Oncol, 2011, 123(3): 581–587.
[30] Schayek H, Bentov I, Jacob-Hirsch J, Yeung C, Khanna C, Helman LJ, Plymate SR, Werner H. Global methylation analysis identifies PITX2 as an upstream regulator of the androgen receptor and IGF-I receptor genes in prostate cancer. Horm Metab Res, 2012, 44(7): 511–519.
[31] Sullivan L, Murphy TM, Barrett C, Loftus B, Thornhill J, Lawler M, Hollywood D, Lynch T, Perry AS. IGFBP7 promoter methylation and gene expression analysis in prostate cancer. J Urol, 2012, 188(4): 1354–1360.
[32] Kanwal R, Pandey M, Bhaskaran N, Maclennan GT, Fu PF, Ponsky LE, Gupta S. Protection against oxidative DNA damage and stress in human prostate by glutathione S-transferase P1. Mol Carcinog, 2014, 53(1): 8–18.
[33] Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, Melnick AM, Elemento O, Demichelis F, Rubin MA. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res, 2013, 73(3): 1232–1244.
[34] Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS, Chang I, Yamamura S, Tanaka Y, Chiyomaru T, Deng GR, Dahiya R. miRNA-34b inhibits prostate cancerthrough demethylation, active chromatin modifications, and AKT pathways. Clin Cancer Res, 2013, 19(1): 73–84.
[35] Majid S, Dar AA, Saini S, Arora S, Shahryari V, Zaman MS, Chang I, Yamamura S, Tanaka Y, Deng G, Dahiya R. miR-23b represses proto-oncogene Src kinase and functions as methylation-silenced tumor suppressor with diagnostic and prognostic significance in prostate cancer. Cancer Res, 2012, 72(24): 6435–6446.
[36] Hulf T, Sibbritt T, Wiklund ED, Patterson K, Song JZ, Stirzaker C, Qu W, Nair S, Horvath LG, Armstrong NJ, Kench JG, Sutherland RL, Clark SJ. Epigenetic-induced repression of microRNA-205 is associated with MED1 activation and a poorer prognosis in localized prostate cancer. Oncogene, 2013, 32(23): 2891–2899.
[37] Li YW, Kong DJ, Ahmad A, Bao B, Dyson G, Sarkar FH. Epigenetic deregulation of miR-29a and miR-1256 by isoflavone contributes to the inhibition of prostate cancer cell growth and invasion. Epigenetics, 2012, 7(8): 940–949.
[38] Shi XB, Xue L, Ma AH, Tepper CG, Gandour-Edwards R, Kung HJ, Devere White RW. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene, 2013, 32(35): 4130–4138.
[39] Borno ST, Fischer A, Kerick M, Falth M, Laible M, Brase JC, Kuner R, Dahl A, Grimm C, Sayanjali B, Isau M, Rohr C, Wunderlich A, Timmermann B, Claus R, Plass C, Graefen M, Simon R, Demichelis F, Rubin MA, Sauter G, Schlomm T, Sultmann H, Lehrach H, Schweiger MR. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation. Cancer Discov, 2012, 2(11): 1024–1035.
[40] Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, Croce N, Vandesompele J, Mestdagh P, Finazzi-Agro E, Levine AJ, Melino G, Bernardini S, Candi E. DNA methylation silences miR-132 in prostate cancer. Oncogene, 2013, 32(1): 127–134.
[41] Suh SO, Chen Y, Zaman MS, Hirata H, Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng GR, Tanaka Y, Dahiya R. MicroRNA-145 is regulated by DNA methylation and p53 gene mutation in prostate cancer. Carcinogenesis, 2011, 32(5): 772–778.
[42] Desotelle J, Truong M, Ewald J, Weeratunga P, Yang B, Huang W, Jarrard D. CpG island hypermethylation frequently silences FILIP1L isoform 2 expression in prostate cancer. J Urol, 2013, 189(1): 329–335.
[43] Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, Huang HD, Lu YY, Teng YC, Lin ST, Lin RK, Tang FM, Lee SB, Hsu HM, Yu JC, Hsiao PW, Juan LJ. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep, 2012, 2(3): 568–579.
[44] Ummanni R, Jost E, Braig M, Lohmann F, Mundt F, Barett C, Schlomm T, Sauter G, Senff T, Bokemeyer C, Sultmann H, Meyer-Schwesinger C, Brummendorf TH, Balabanov S. Ubiquitin carboxyl-terminal hydrolase 1 (UCHL1) is a potential tumour suppressor in prostate cancer and is frequently silenced by promoter methylation. Mol Cancer, 2011, 10: 129.
[45] Olkhov-Mitsel E, Van Der Kwast T, Kron KJ, Ozcelik H, Briollais L, Massey C, Recker F, Kwiatkowski M, Fleshner NE, Diamandis EP, Zlotta AR, Bapat B. Quantitative DNA methylation analysis of genes coding for kallikrein-related peptidases 6 and 10 as biomarkers for prostate cancer. Epigenetics, 2012, 7(9): 1037–1045.
[46] Kim YJ, Yoon HY, Kim SK, Kim YW, Kim EJ, Kim IY, Kim WJ. EFEMP1 as a novel DNA methylation marker for prostate cancer: array-based DNA methylation and expression profiling. Clin Cancer Res, 2011, 17(13): 4523–4530.
[47] Vinarskaja A, Goering W, Ingenwerth M, Schulz WA. ID4 is frequently downregulated and partially hypermethylated in prostate cancer. World J Urol, 2012, 30(3): 319–325.
[48] Gao TY, He BS, Pan YQ, Li R, Xu YQ, Chen LP, Nie ZL, Gu L, Wang SK. The association of retinoic acid receptor beta2(RARbeta2) methylation status and prostate cancer risk: a systematic review and meta-analysis. PLoS ONE, 2013, 8(5): e62950.
[49] Dumache R, Puiu M, Minciu R, Bardan R, David D, Tudor A, Bumbacila B. Retinoic acid receptor beta2 (RARbeta2): nonivasive biomarker for distinguishing malignant versus benign prostate lesions from bodily fluids. Chirurgia (Bucur), 2012, 107(6): 780–784.
[50] Yoon HY, Kim YW, Kang HW, Kim WT, Yun SJ, Lee SC, Kim WJ, Kim YJ. Pyrosequencing analysis of APC methylation level in human prostate tissues: A molecular marker for prostate cancer. Korean J Urol, 2013, 54(3): 194–198.
[51] Sharma P, Chinaranagari S, Patel D, Carey J, Chaudhary J. Epigenetic inactivation of inhibitor of differentiation 4 (Id4) correlates with prostate cancer. Cancer Med, 2012, 1(2): 176–186.
[52] Yaqinuddin A, Qureshi SA, Pervez S, Bashir MU, Nazir R, Abbas F. Frequent DNA hypermethylation at the RASSF1A and APC gene loci in prostate cancer patients of pakistani origin. ISRN Urol, 2013, 627249.
[53] 刘岗, 殷波, 宋永胜. 前列腺癌组织 RASSF2基因甲基化及蛋白表达的检测及意义. 中华男科学杂志, 2013, 19(2): 107–110.
[54] Dansranjavin T, Wagenlehner F, Gattenloehner S, Steger K, Weidner W, Dammann R, Schagdarsurengin U. Epigenetic down regulation of RASSF10 and its possible clinical implication in prostate carcinoma. Prostate, 2012, 72(14): 1550–1558.
[55] Truong M, Yang B, Wagner J, Kobayashi Y, Rajamanickam V, Brooks J, Jarrard DF. Even-skipped homeobox 1 is frequently hypermethylated in prostate cancer and predicts PSA recurrence. Br J Cancer, 2012, 107(1): 100–107.
[56] Liu WW, Gong J, Hu J, Hu TH, Sun YF, Du JH, Sun CY, Guan M, Jiang HW, Lu Y. Quantitative assessment of AKAP12 promoter methylation in human prostate cancer using methylation-sensitive high-resolution melting: correlation with Gleason score. Urology, 2011, 77(4): 1006.e1–1006.e7.
[57] Sørensen KD, Abildgaard MO, Haldrup C, Ulhøi BP, Kristensen H, Strand S, Parker C, Høyer S, Borre M, Ørntoft TF. Prognostic significance of aberrantly silenced ANPEP expression in prostate cancer. Br J Cancer, 2013, 108(2): 420–428.
[58] Chiam K, Centenera MM, Butler LM, Tilley WD, Bianco-Miotto T. GSTP1 DNA methylation and expression status is indicative of 5-aza-2'-deoxycytidine efficacy in human prostate cancer cells. PLoS ONE, 2011, 6(9): e25634.
[59] Wu T, Giovannucci E, Welge J, Mallick P, Tang WY, Ho SM. Measurement of GSTP1 promoter methylation in body fluids may complement PSA screening: a meta-analysis. Br J Cancer, 2011, 105(1): 65–73.
[60] Mitsui Y, Shiina H, Hiraki M, Arichi N, Hiraoka T, Sumura M, Honda S, Yasumoto H, Igawa M. Tumor suppressor function of PGP9. 5 is associated with epigenetic regulation in prostate cancer--novel predictor of biochemical recurrence after radical surgery. Cancer Epidemiol Biomarkers Prev, 2012, 21(3): 487–496.
[61] Vasiljević N, Ahmad AS, Beesley C, Thorat MA, Fisher G, Berney DM, Møller H, Yu Y, Lu YJ, Cuzick J, Foster CS, Lorincz AT. Association between DNA methylation of HSPB1 and death in low Gleason score prostate cancer. Prostate Cancer Prostatic Dis, 2013, 16(1): 35–40.
[62] Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene, 2002, 21(35): 5400–5413.
[63] De Smet C, Loriot A. DNA hypomethylation in cancer: Epigenetic scars of a neoplastic journey. Epigenetics, 2010, 5(3):
[64] Yegnasubramanian S, Haffner MC, Zhang YG, Gurel B, Cornish TC, Wu ZJ, Irizarry RA, Morgan J, Hicks J, Deweese TL, Isaacs WB, Bova GS, De Marzo AM, Nelson WG. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res, 2008, 68(21): 8954–8967.
[65] Paradowska A, Fenic I, Konrad L, Sturm K, Wagenlehner F, Weidner W, Steger K. Aberrant epigenetic modifications in the CTCF binding domain of the IGF2/H19 gene in prostate cancer compared with benign prostate hyperplasia. Int J Oncol, 2009, 35(1): 87–96.
[66] Bhusari S, Yang B, Kueck J, Huang W, Jarrard DF. Insulin-like growth factor-2 (IGF2) loss of imprinting marks a field defect within human prostates containing cancer. Prostate, 2011, 71(15): 1621–1630.
[67] Vestergaard EM, Nexø E, Torring N, Borre M, Ørntoft TF, Sørensen KD. Promoter hypomethylation and upregulation of trefoil factors in prostate cancer. Int J Cancer, 2010, 127(8): 1857–1865.
[68] Yoon HY, Kim SK, Kim YW, Kang HW, Lee SC, Ryu KH, Shon HS, Kim WJ, Kim YJ. Combined hypermethylation of APC and GSTP1 as a molecular marker for prostate cancer: quantitative pyrosequencing analysis. J Biomol Screen, 2012, 17(7): 987–992.
[69] Van Neste L, Herman JG, Otto G, Bigley JW, Epstein JI, Van Criekinge W. The epigenetic promise for prostate cancer diagnosis. Prostate, 2012, 72(11): 1248–1261.
[70] Trock BJ, Brotzman MJ, Mangold LA, Bigley JW, Epstein JI, Mcleod D, Klein EA, Jones JS, Wang S, Mcaskill T, Mehrotra J, Raghavan B, Partin AW. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU Int, 2012, 110(1): 56–62.
[71] Truong M, Yang B, Livermore A, Wagner J, Weeratunga P, Huang W, Dhir R, Nelson J, Lin DW, Jarrard DF. Using the epigenetic field defect to detect prostate cancer in biopsy negative patients. J Urol, 2013, 189(6): 2335–2341.
[72] Sabatino MA, Geroni C, Ganzinelli M, Ceruti R, Broggini M. Zebularine partially reverses GST methylation in prostate cancer cells and restores sensitivity to the DNA minor groove binder brostallicin. Epigenetics, 2013, 8(6): 656–665.
[73] Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer, 2010, 126(11): 2520–2533.
[74] Agarwal S, Amin KS, Jagadeesh S, Baishay G, Rao PG, Barua NC, Bhattacharya S, Banerjee PP. Mahanine restoresRASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol Cancer, 2013, 12(1): 99.
[75] Weng JR, Lai IL, Yang HC, Lin CN, Bai LY. Identification of Kazinol Q, a Natural Product from Formosan Plants, as an Inhibitor of DNA Methyltransferase. Phytother Res, 2014, 28(1): 49–54.
[76] Lin JQ, Haffner MC, Zhang YG, Lee BH, Brennen WN, Britton J, Kachhap SK, Shim JS, Liu JO, Nelson WG, Yegnasubramanian S, Carducci MA. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate, 2011, 71(4): 333–343.
[77] Graca I, Sousa E, Baptista T, Almeida M, Ramalho-Carvalho J, Palmeira C, Henrique R, Jerónimo C. Antitumoral effect of the non-nucleoside DNMT inhibitor RG108 in human prostate cancer cells. Curr Pharm Des, 2013.
[78] Rabiau N, Trraf HK, Adjakly M, Bosviel R, Guy L, Fontana L, Bignon YJ, Bernard-Gallon DJ. miRNAs differentially expressed in prostate cancer cell lines after soy treatment. In Vivo, 2011, 25(6): 917–921.
[79] Adjakly M, Bosviel R, Rabiau N, Boiteux JP, Bignon YJ, Guy L, Bernard-Gallon D. DNA methylation and soy phytoestrogens: quantitative study in DU-145 and PC-3 human prostate cancer cell lines. Epigenomics, 2011, 3(6): 795–803.
[80] Khor TO, Huang Y, Wu TY, Shu LM, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol, 2011, 82(9): 1073–1078.
[81] Liu ZW, Ren GL, Shangguan CY, Guo LJ, Dong ZX, Li YY, Zhang WN, Zhao L, Hou PF, Zhang Y, Wang XL, Lu J, Huang BQ. ATRA inhibits the proliferation of DU145 prostate cancer cells through reducing the methylation level of HOXB13 gene. PLoS ONE, 2012, 7(7): e40943.
[82] Dietrich D, Hasinger O, Bañez LL, Sun L, Van Leenders GJ, Wheeler TM, Bangma CH, Wernert N, Perner S, Freedland SJ, Corman JM, Ittmann MM, Lark AL, Madden JF, Hartmann A, Schatz P, Kristiansen G. Development and clinical validation of a real-time PCR assay for PITX2 DNA methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. J Mol Diagn, 2013, 15(2): 270–279.
(责任编委: 朱卫国)
DNA methylation of prostate cancer and clinical application
Fan Zhao, Ze Yang
The 5th Medical College of Peking University, Institute of Geriatrics, Chinese Ministry of Health, Beijing Hospital, Beijing 100730, China
It is well-known that the interation of both genetic and epigenetic mechanisms results in the formation of malignant tumor. Epigenetic mechanism includes DNA methylation, histone modifications, and miRNA regulation. DNA aberrant methylation (hypermethylation and hypomethylation), which leads to genomic instability and inappropriate gene expression, is the best-characterized alteration in prostate cancer. It plays an important role in the initiation and development of prostate cancer. Meanwhile, DNA methylation, as a hotspot in researches of epigenetics, would provide a new methodology and approach for early clinical diagnosis, prognosis and medication treatment of prostate cancer. According to recent studies on DNA hypermethylation and DNA hypomethylation, this review highlights the potential epigenetic mechanism of prostate cancer and discusses the latest research progress in clinical translation.
prostate cancer; DNA hypermethylation; DNA hypomethylation
2013-12-09;
2014-01-14
国家自然科学基金项目(编号:30972709, 81061120527, 81241082), 北京医院重大基金项目(编号:BJ-2010-30), 卫生部部属医院临床学科重点项目(编号:01020101), 卫生部行业基金项目(编号:201302008)和科技部十二五支撑计划项目(编号:2012BAI10B01)资助
赵帆, 硕士研究生, 专业方向:医学遗传学。E-mail: zhaofan1219@163.com
杨泽, 研究员, 博士生导师, 研究方向:医学遗传学。E-mail: yang_ze@sina.com
10.3724/SP.J.1005.2014.0420
时间: 2014-3-20 14:48:59
URL: http://www.cnki.net/kcms/detail/11.1913.R.20140320.1448.001.html

