Deletion of the waaf gene affects O antigen synthesis and pathogenicity in Vibrio parahaemolyticus from shell fish
Feng Zho, Guoying Ding, Qilong Wng, Huihui Du, Guosheng Xio,*, Deqing Zhou*
a College of biology and food engineering, Chongqing Three Gorges University, Wanzhou 404100, China
b Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Laboratory for Marine Drugs and Bioproducts of Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266071, China
Keywords:
Vibrio parahaemolyticus
waaf gene
LPS
O antigen
Pathogenicity
A B S T R A C T
Vibrio parahaemolyticus is the main cause of foodborne gastroenteritis, which is widely distributed in shell fish and other seafood. Most V. parahaemolyticus are nonpathogenic, and only a few types, such as serotype O3:K6, are pathogenic, which is also the most prevalent strain in Asia. However, the relationship between this serotype and pathogenicity has yet to be established. The waaf gene is located in the O antigen synthesis gene cluster. Thus, we constructed a waaf gene deletion mutant (i.e., △waaf) of wild-type (WT) which isolated from shell fish serotype O3:K6 via chitin-mediated transformation technology. We then constructed the △waaf complementary strain (i.e., C-Δwaaf) via the Escherichia coli S17 λpir strain by conjugation. The basic physiological characteristics, adhesion to Caco2 cells, and pathogenicity of the WT, Δwaaf, and C-Δwaaf strains were compared. Growth curves showed no remarkable differences between the WT and Δwaaf strains.However, the Δwaaf strain non-reactive to O3 antisera and other 12 O-group antisera of V. parahaemolyticus.Moreover, the number of flagella and extracellular polysaccharides decreased, the adhesion decreased, and the pathogenicity weakened. These characteristics of the C-Δwaaf strain were similar to those of the WT strain. These results indicated that the waaf gene is vital to the serotype in V. parahaemolyticus, and changes in O antigen could affect the pathogenicity of this bacterium. This study will be helpful to understand the pathogenic mechanism of V. parahaemolyticus.
1. Introduction
Vibrioparahaemolyticusis a Gram-negative halophilic bacterium that is widely distributed in marine and estuarine environments [1,2].Humans usually suffer from gastroenteritis due toV.parahaemolyticusby eating raw or poorly heated seafood. In severe cases, septicemia may occur [3]. The typical clinical phenomena ofV.parahaemolyticusinfection are acute dysentery and abdominal pain, accompanied by diarrhea, nausea, vomiting, low fever, chills,and watery stool [4]. Since 1996, outbreaks of food poisoning due toV.parahaemolyticusare mainly concentrated in a few serotypes(e.g., O3:K6, O4:K68 and O1:KUT) that constitute the main epidemic group ofV.parahaemolyticus[5,6]. Among them, the O3:K6 type accounts for about 80% of pathogenicV.parahaemolyticusand has a very high infection and transmission capacity [7,8]. Some specific serotypes may be more relevant to human diseases [9,10]. The O3:K6 epidemic groups may have evolved from the same ancestor through recombination of antigen gene cluster and its adjacent sequences [11],but the exact evolutionary process, pathogenic mechanism, and genetic basis of serotype diversity ofV.parahaemolyticusremain unclear. The global outbreak ofV.parahaemolyticuswarrants the investigation of its virulence factors and its impact on human hosts.
Thermostable direct hemolysin (TDH), thermostable related hemolysin (TRH), thermolabile hemolysin (TLH) and type III secretion system (T3SS) are the main pathogenic factors ofV.parahaemolyticus, TDH is usually contained in pathogenic O3:K6 serotype [12,13]. Two sets of T3SS, T3SS1 and T3SS2 were found inV.parahaemolyticus, which were located on chromosome 1 and 2 respectively. T3SS1 is related to the environmental adaptability,and T3SS2 is mainly involved in the enterotoxicity [13]. There are more than 80 serum combination of O-group and K-type inV.parahaemolyticus, but only 13 O-group and 71 K-type antigens have been commercialized [14]. There are still many new and unidentified serotypes ofV.parahaemolyticus. Lipopolysaccharide(LPS, O antigen) and capsular polysaccharide (CPS, K antigen)provide a basis for serotyping [15,16]. As a unique structure of Gramnegative bacteria, LPS plays an important role in the pathogenesis of most Gram-negative pathogens [17]. A typical LPS consists of three parts: O-specific polysaccharide side chain, lipid A, and core oligosaccharide [18].V.parahaemolyticuslacks O-specific polysaccharide side chain, and its O antigen phenotype is determined by the core oligosaccharide, an important component of LPS [19].O antigen synthesis genes usually appear in clusters, which are called O antigen gene clusters. Complete genome sequencing ofV.parahaemolyticushas been completed, which shows that the region A located on chromosome 1 is supposed to be related to O antigen synthesis [20,21], thereby laying a theoretical foundation for screening and identifying the key genes ofV.parahaemolyticusO antigen synthesis. The O antigen gene cluster of Gram-negative bacteria usually contains three kinds of genes: monosaccharide synthesis genes, glycosyl transfer genes, and oligosaccharide unit treatment genes. Thewaaorrfagene family is responsible for core oligosaccharide synthesis. This gene family is common in Gramnegative bacteria [22]. Thewaagene family is responsible for assembling the core oligosaccharide of LPS inEscherichia coli[23].In other genera, such asSalmonellaandKlebsiella pneumonia, thewaagene family is also included in the core polysaccharide synthesis gene cluster [24,25]. VP0212 (the putativewaafgene) is located on region A in chromosome 1 ofV.parahaemolyticus.waafis a gene encoding heptyltransferase II, an important enzyme for the synthesis of core oligosaccharides [26]. The loss of its function leads to changes in the structure of core oligosaccharides, which in turn could lead to changes in the structure of O antigen in Gram-negative bacteria.
In this study, thewaafgene was knocked out fromV.parahaemolyticusisolates from shell fish serotype O3:K6. Δwaafstrain and complementary strain C-Δwaafwere constructed to verify the role of this gene in O antigen synthesis. This study aimed to explore the relationship between serotype and pathogenicity ofV.parahaemolyticus.
2. Materials and methods
2.1 Bacterial strains
Bacterial strains and plasmids used in this study are shown in Table 1.E. colistrain was cultivated on Luria-Bertani (LB, Land Bridge Technology, Beijing, China) or a solid medium containing 1.5% agar at 37 °C with or without shaking.V.parahaemolyticusstrain was cultivated in MLB medium (Luria-Bertani medium supplemented with 3% NaCl) and incubated at 37 °C. If necessary, the medium was supplemented with appropriate reagent and antibiotics,namely 50 µg/mL Chloramphenicol (Cm) and 50 µg/mL Kanamycin(Kan).E. coliS17 λpir and plasmid pBBR1MCS-2 were used for making the complementary strain.
Table 1Bacterial strains and plasmids used in this study.
2.2 Experimental animal
The 4–5 week SPF BALBC mice were purchased from Drug Test Institute of Chongqing. The 3R principle of experimental animals was observed in the experiment and all animal experiments have been approved by the ethics committee of Chongqing Three Gorges University.
2.3 Construction of homologous recombination fragment
The homologous recombination fragment was constructed fromV.parahaemolyticusWT strain by using the oligonucleotide primers listed in Table 2. According to the whole genome sequence ofV.parahaemolyticus[20], the upstream and downstream homologous arms ofwaafgene were amplified via PCR by using the WT strain chromosomal DNA as a template and the primer pairswaaf-U-F/waaf-U-R andwaaf-D-F andwaaf-D-R. The Cm resistance gene was amplified using the pKD3 vector as a template and the primer pairs PKD3-F/PKD3-R. Both ends of this amplified fragment had the complementary sequence of 3’ upstream homologous arm and 5’ downstream homologous arm. The three fragments were fused in the order of upstream homologous arm, Cm-resistant gene, and downstream homologous arm to form a homologous recombinant fragment via fusion PCR [27]. The homologous recombinant fragment was used as templates in a second PCR by using the primerswaaf-U-F/waaf-D-R. The amplicon was purified and cloned into pMD18-T vector, and then the recombinant plasmid was introduced into TOP 10 strain. The integrity and accuracy of the recombinant plasmid was verified by sequencing.

Table 2Primers used in this paper.
2.4 Chitin-mediated transformation
Thewaafgene deletion mutant was constructed via chitinmediated transformation.
The WT strain was inoculated in MLB and shaken at 150 r/min at 37 °C for 6 h. The aforementioned cultures were diluted at a ratio of 1:100 into fresh MLB and grown with shaking at 150 r/min at 37 °C until OD600nm= 0.5. Afterward, 1 mL of the bacterium liquid was collected and placed into a tube and then centrifuged at 5 000 r/min for 5 min to precipitate the bacteria. The bacteria were resuspended by sterilized sea water to make OD600nm= 0.2.
Chitin-base natural transformation ofV.parahaemolyticuswas performed according to previous method [28]with minor modifications. A piece of 1 cm2sterilized crab shell was put into a 12-well cell culture plate (Corning, New York, USA) with 2 mL bacterial suspension, and then 2-4 µg of fusion DNA fragment was added into the well and cultured overnight at 30 °C. The following day,the crab shells were removed from the well and washed once with phosphate-buffered saline, placed into a 50 mL tube containing 2 mL of phosphate-buffered saline, and vortexed to release bacteria. The supernatant was diluted and plated with 5 µg/mL of Cm resistance and cultured for 18-24 h until single colonies were grown. A single colony was selected and inoculated into a 2 mL tube added with 1 mL MLB with 5 µg/mL Cm and grown with shaking at 150 r/min at 37 °C for about 6 h. The bacterial solution was then collected for PCR verification.
2.5 Construction of genetic complementary strain
The complementary strain of deletion mutant was constructed by amplifying thewaafgene from WT strain genomic DNA via PCR by using the primer pairs HB-S-KpnI/HB-X-XbaI. The PCR products were purified and digested by restriction endonucleasesKpnI andXbaI,and then the fragment was transformed into pBBR1MCS-2 vector.This recombinant plasmid (pBBR1MCS-2-waaf) was introduced intoE.coliS17 λpir and then was mated with △waafaccording to previous methods [29]. The complementary strain (C-Δwaaf) was obtained by screening with TCBS agar plates containing 5 µg/mL Kan. A single colony was confirmed via PCR and then stored for later use.
2.6 Serologic testing
Serotyping was done using a commercially availableV.parahaemolyticusantisera test kit (Denka Seiken, Tokyo, Japan).The fresh cultures grown on MLB plate were used for serological test. O grouping and K typing are carried out by slide agglutination following the manufacturer’s instructions.
2.7 Bacterial growth curve assays
Subcultures of the strains were incubated overnight at 37 °C, diluted in MLB at a ratio of 1:600, inoculated in a flask, and incubated at 37 °C at 150 r/min. OD600nmvalue was measured every 2 h until the bacteria entered the plateau stage, and then the growth curve was drawn.
2.8 Adhesion assays
The assays of adhesion to Caco2 were performed according to previous methods [30]with minor modifications. In brief, each bacterial strain was washed and resuspended in approximately 2 × 107CFU/mL, added to confluent Caco2 cultured in a 24-well plate at a multiplicity of infection of 10. The plates at 37 °C in 5% CO2were incubated for 60 min to allow microbe-cell interactions.Nonspecific bacterial adhesion was removed by vigorously washing the cells three times and then lysing them by incubation in 0.5% Triton X-100. Appropriate dilutions were plated to obtain total CFU.
2.9 Real-time fluorescence quantitative RT-PCR (qPCR)
Bacterium RNA was extracted as previously described [31]via qPCR following the instructions of SYBR premix ExTaqTM II kit. Each PCR was performed with a 25 μL final volume containing 2 μL of cDNA, 3.5 mmol/L MgCl2, 200 μmol/L deoxynucleoside triphosphate, 300 nmol/L of each primer, and 1.25 U ofTaqDNA polymerase. The thermal cycling conditions were as follows: predenaturation at 95 °C for 30 s, 40 cycles of denaturation at 95 °C for 5 s,and then annealing and extension at 60 °C for 30 s. All PCRs were run on an ABI Viia7 Real-time PCR System, and each experiment was performed with three technical replicates. The oligonucleotide primers were listed in Table 2. The relative expression of genes was calculated via the 2-ΔΔCTmethod [32].
2.10 Challenge experiments in mice
Sixty mice were randomly divided into 4 groups. Each group was fed in one cage. They were fasted but allowed to drink freely for 12 h before the experiment. During the experiment, they were fed normally. WT, Δwaaf, and C-Δwaafstrains were inoculated in MLB and shaken at 160 r/min at 37 °C for 10–16 h. The bacteria were centrifuged at 5 000 r/min, diluted with sterilized PBS solution to about 5 × 108CFU/mL. Female SPF BALBC mice were gavaged with 0.5 mL bacterial solution, whereas the control group was given with 0.5 mL PBS solution. The mice were observed at 2, 4, 6 and 12 h after gavage and then observed every 12 h for 96 h. The state of mice and the number of dead mice were recorded, and the survival rate curve was drawn.
2.11 Histological examination and intestinal bacterial load
The visceral lesions of the mice that died after the challenge were observed. The liver and small intestines of the mice were fixed with formaldehyde, dehydrated with ethanol, embedded in paraffin,sliced and stained with eosin, and then observed under a microscope.To calculate intestinal bacterial load, the intestinal tissues of mice infected withV.parahaemolyticuswere homogenized, gradient diluted, and then spread on MLB plate for counting.
2.12 Kanagawa phenomenon (KP) hemolytic test
Colonies ready from TCBS (Land Bridge Technology, Beijing,China) plates were used for Kanagawa phenomenon (KP) test. They were streaked onto Wagatsuma blood agar plates that contained 7% NaCl, 3 g yeast extract, 10 g peptone, 15 g agar, 5 g K2HPO4, 10 g mannitol, 0.001 g crystal violet, 1 L distilled water, and 20% rabbit defibrinated blood. The plates were then incubated for 18–24 h at 37 °C. A well-defined, hemolysis zone around and under a single colony was recorded as KP positive [33].
2.13 Transmission electron microscopy
Under aseptic conditions, the WT and Δwaafstrains were streakinoculated onto MLB solid medium and incubated at 37 °C for 7–8 h. A single colony was washed with 1 × PBS buffer solution(pH 7.2) three times and then gently mixed well. The bacteria were stained by 2% osmium acid with equal volume, and then bacterial samples were placed on grids. Excess liquid was absorbed using a filter paper. The samples were dried and loaded for transmission electron microscopy.
2.14 Statistical analysis
Tukey test with analysis of variance (ANOVA) was performed to evaluate the statistical significant of the findings.P< 0.05 were considered to be statistically significant (*), whileP< 0.01 were considered as extremely significant (**).
3. Results and discussion
3.1 Construction and confirmation of Δwaaf and C-Δwaaf strains
The homologous recombination fragment of the construction process is shown in Fig. S1. The upstream and downstream homologous arms were amplified from the WT strain with a size of 1 230 and 1 143 bp, respectively (Fig. S1A). The Cm-resistant gene of 1 053 bp was amplified from the pKD3 vector (Fig. S1A).The homologous recombination fragment included an upstream homologous arm, a Cm-resistant gene, and a downstream homologous arm with a size of 3 386 bp (Fig. S1B). The confirmation of Δwaafafter chitin-mediated transformation is shown in Fig. S1C. The 1 059 bp fragment, which is the full length of thewaafgene, could not be amplified from the Δwaafgenome by the primer pairwaafn-F andwaaf-n-R. By contrast, the 1 053 bp fragment, which is the Cm-resistant gene, was successfully amplified from the genome(Fig. S1C, Lanes 1 and 3). Thetlhgene, which is a genus-specific gene, and thetdhgene, which is the pathogenic characteristic gene ofV.parahaemolyticus, were successfully amplified from the Δwaafgenome with a size of 500 and 269 bp, respectively (Fig. S1C, Lanes 4 and 5). These results demonstrated that the full length of thewaafgenes was knocked out in the Δwaafstrain.
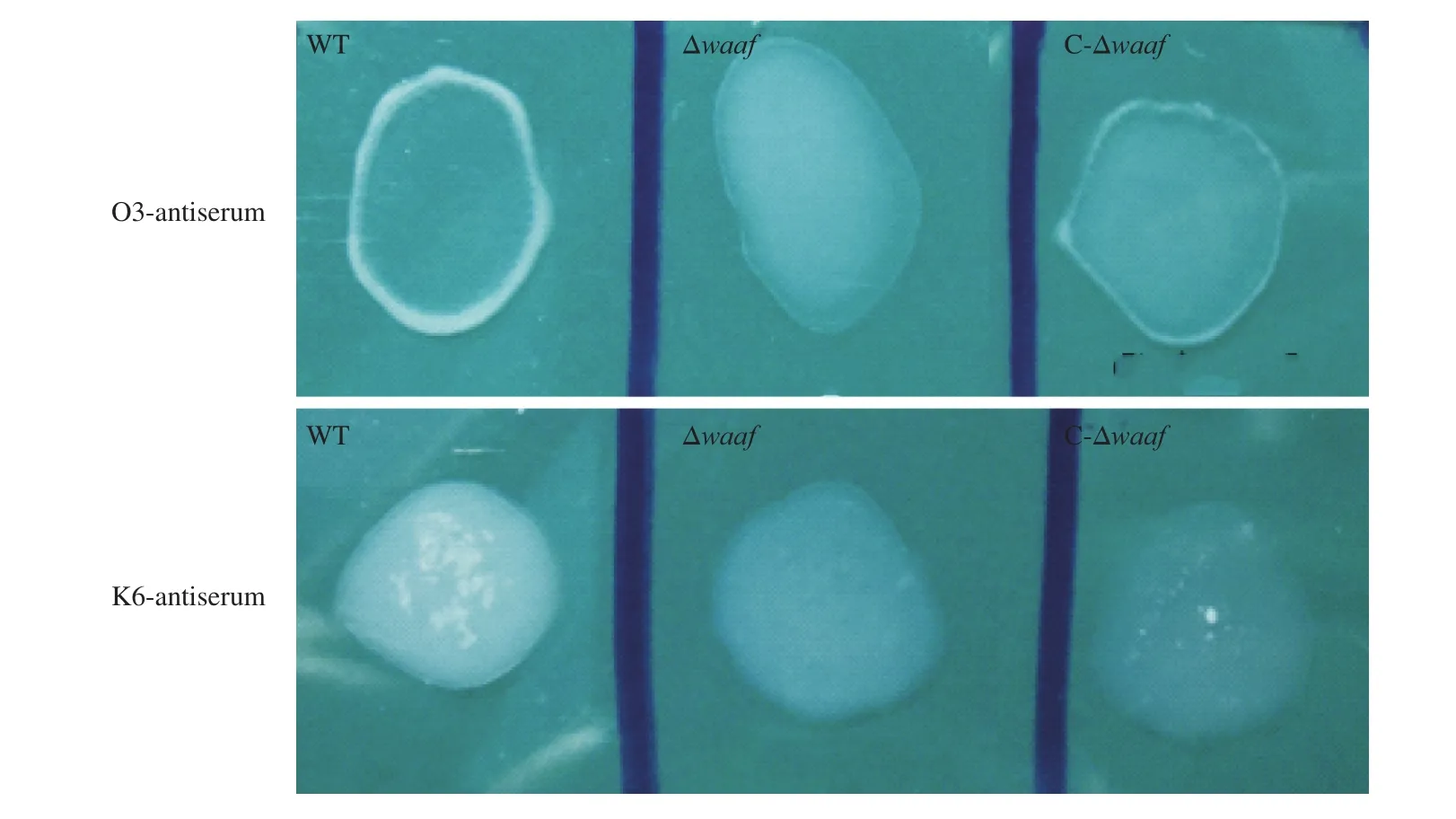
Fig. 1 The serotyping of WT, Δwaaf and C-Δwaaf strains. Agglutination of different bacteria with (A) O3-antiserum and (B) K6-antiserum. There were obvious flocculation occurred in WT and C-Δwaaf strains within 1 min, but there was no flocculation occurred in Δwaaf strains.
The function of thewaafgene was further verified by constructing the genetic complementary strain of Δwaaf(C-Δwaaf) by usingE. coliS17λpir. The results of confirmation are shown in Fig. S2. In the C-Δwaafstrain, the full length of thewaafgene was successfully amplified (Fig. S2, Lane 1), and thetlhandtdhgenes were also successfully amplified from the genome (Fig. S2, Lanes 2 and 3).Results showed that thewaafgene was re-transformed into the Δwaafstrain, and the C-Δwaafstrain was successfully constructed.
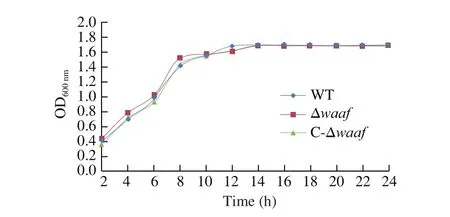
Fig. 2 The growth curves of different strains. The WT, Δwaaf and C-Δwaaf strains were incubated in MLB. In 24 h, there was no significant difference among the three strains by ANOVA analysis.
3.2 Basic biological characteristics of the Δwaaf and C-Δwaaf strains
3.2.1 Serotyping
The WT strain was isolated from patients with diarrhea, and its serotype was O3:K6. When thewaafgene was knocked out, the activity of O antigen disappeared in Δwaafstrain, and it did not agglutinate with the O3 antiserum and other 12 O-group antisera ofV.parahaemolyticus. When thewaafgene was re-transformed into the Δwaafstrain, the C-Δwaafstrain coagulated with the O3 antiserum as quickly as the WT strain (Fig. 1). These results indicated that thewaafgene is a key gene regulating the serotype ofV.parahaemolyticus.The presence or absence of thewaafgene directly affects the serotype of this bacterium.
In most Gram-negative bacteria, the O antigen is determined by the specific polysaccharide side chain. However, the O polysaccharide side chain is absent inV.parahaemolyticus, and its O antigen characteristics are determined by the core polysaccharide [34].Heptose is an important component of core polysaccharide.waa(also known asrfa) gene clusters are responsible for connecting heptose I (Hep I), heptose II (Hep II) and heptose III (Hep III) to the 3-deoxy-d-mannan octyl glycolic acid (KDO) structure on the core polysaccharide [35]. In most Gram-negative bacteria, the absence of heptanyl results in serious shortening of the glycochain of LPS and the disappearance of O antigen characteristics.
The K type ofV.parahaemolyticusis determined by CPS. The capsule composed of CPS is an extracellular structure of bacteria. The biosynthesis pathway of CPS is the ABC pathway or the wzx/wzy pathway [36,37]. Genes related to CPS biosynthesis always exist as gene clusters in the genome, which are called CPS gene clusters or K antigen gene clusters. InV.parahaemolyticus, the K antigen gene cluster is located between thegmhdgene and therjggene in chromosome 1 and is closely adjacent to the O antigen gene cluster [38]. When thewaafgene was knocked out, the mutant did not agglutinate with K type antiserum.These results showed that the deletion ofwaafgene affects not only the O antigen but also the K antigen.
3.2.2 Growth curves of different strains
The growth curves of the WT, Δwaaf, and C-Δwaafstrains are plotted in Fig. 2. There was no significant difference in growth rate among the three strains (P> 0.05), indicating that the deletion ofwaafgene does not affect cell growth.
3.2.3 Transmission electron micrographs
LPS, CPS, and flagella are important components of bacterial surface [39]. Compared with the surface of the WT strain, the surface of the Δwaafstrain remarkably changed (Fig. 3). More flagella were observed around the cells of WT strains but not around the cells of the Δwaafstrain. More extracellular polysaccharides of mucilage layer were observed around the cells of WT strains, whereas no such substances were observed around the cells of the Δwaafstrain. In addition, separating the WT cells during the preparation process before transmission electron microscopy was more difficult,indicating that extracellular polysaccharides play an important role in cell adhesion. Preliminary observations indicated that the Δwaafstrain might affect the secretion of extracellular polysaccharides and the synthesis of flagellum.
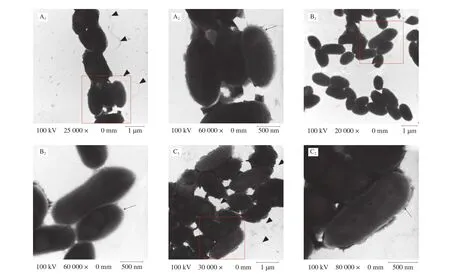
Fig. 3 Transmission electron microscope images. A1, A2 were WT strain; B1, B2 were Δwaaf strain; C1, C2 were C-Δwaaf strain. A2, B2 and C2 were partial enlargement of the position indicated with red frame (A1, B1 and C1). Triangular indicate the flagellum, and arrows indicate the LPS/capsule layer. The mutant strain shows a significant decrease in the heavily stained dense layer of polysaccharide that surrounds the WT strain.
Both LPS and flagella influence the characteristics of cell membranes, such as hydrophobicity, automatic aggregation, antigenic properties, and biofilm formation. LPS and flagellum are highly specific and variable, and they can also activate host immune activity.InE. coli, the structural modification of LPS affects bacterial cell motility, which is closely related to bacterial flagella. The rotation of the flagellum promotes the release of LPS from the intracellular matrix into the flagellum. The close relationship between LPS and flagella was also found inV. fischeriandV. cholera[1,40]. LPS and flagellum influence each other, but their relationship remains unclear.In this study, the same phenomenon was observed. Thewaafgene was located in the gene cluster of O antigen synthesis, and its deletion led to the disappearance ofV.parahaemolyticusO antigen characteristics.Moreover, the deletion of thewaafgene affected the synthesis of flagella, reduced the number of flagella, and affected the secretion of polysaccharides.
3.3 Adhesion
Adhesion is the key processes to bacterial infection. The main target ofV.parahaemolyticusis the intestinal mucosal cells of its host. In this study, Caco2 cells were used to simulate the adhesion of this bacterium. The adhesion of the Δwaafstrain were substantially lower than those of the WT strain and the complementary strain(Fig. 4), indicating that the adhesion ofV.parahaemolyticusto its host are related to thewaafgene.
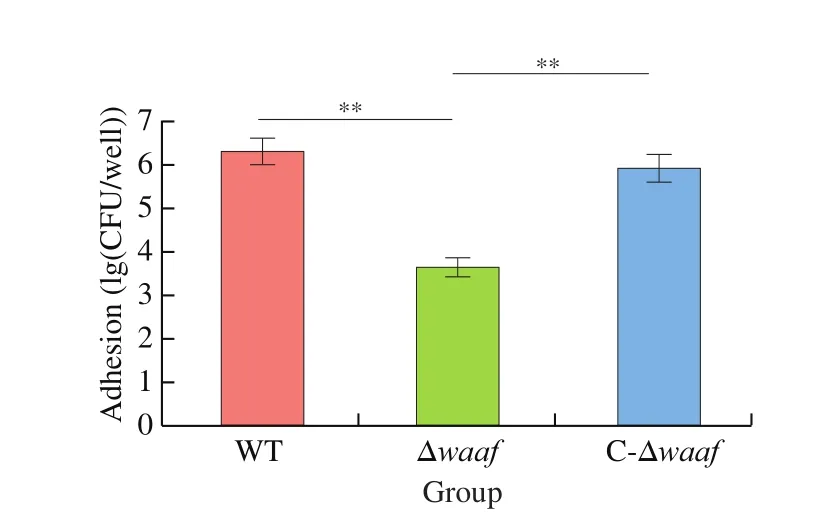
Fig. 4 Adhesion of WT, Δwaaf and C-Δwaaf strains. Comparison of adhesion among WT, Δwaaf and C-Δwaaf strains, the number of V. parahaemolyticus were obtained by colony count, and the log of this number were used to evaluate adhesion of different strains in Caco2 cells.** indicate there is a significant difference between them (P < 0.01).
3.4 Expression of the tdh gene and upstream and downstream genes of waaf
Results of serotyping, transmission electron microscopy, adhesion assays revealed that the deletion of thewaafgene can influence the basic biological characteristics ofV.parahaemolyticus. Does the deletion affect other aspects at the transcriptional level? Thus, we selected thetdhgene, which is the important virulence factor and upstream and downstream genes ofwaaf, to further analyze their mRNA expression via qPCR. Only the downstream gene (VP0211)was downregulated in the Δwaafstrain compared with those in the WT strain. The expression levels of thetdhgene and the downstream gene (VP0213) were the same as those of the WT strain (Fig. 5).
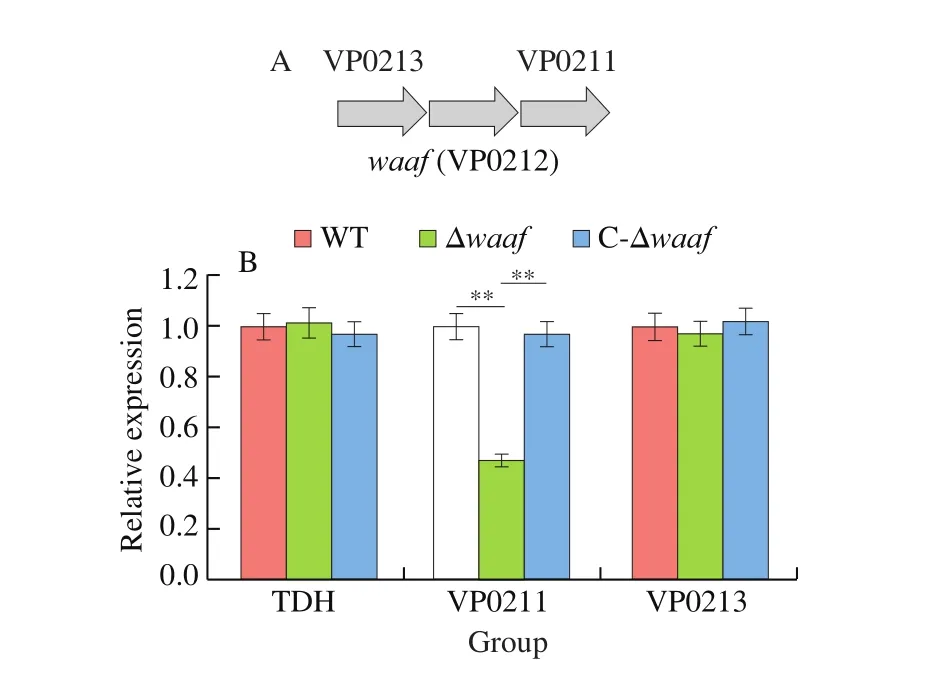
Fig. 5 The expression of tdh gene and upstream and downstream genes by qPCR. ** indicate there is a significant difference between them (P < 0.01).
3.5 Pathogenicity
3.5.1 Hemolytic characteristics
KP hemolysis test is a classic experiment to determine the virulence ofV.parahaemolyticus. Most pathogenicV.parahaemolyticusstrains contain thetdhgene, which can produce direct thermostable hemolysis (TDH). TDH can produce hemolytic zones on Wagatsuma blood agar plates. Environmental isolates usually do not contain TDH, so hemolysis will not occur. When the WT, Δwaaf, C-Δwaaf, and control strains (1.199 7,tdh-,trh+) were inoculated onto the blood agar plates, except for the control strain, the three strains all produced hemolytic zones. These results, combined with those of qPCR, demonstrated that the deletion of thewaafgene does not affect the expression and function of thetdhgene.
3.5.2 Infection experiments in mice
The pathogenicity of the WT, Δwaaf, and C-Δwaafstrains was compared via gavage by using a murine infection model. In the WT strain group, 2 h after gavage with the strains, some mice showed listlessness, eating less, decrease in exercise, accumulation in piles,and bristles and tremors of the whole body. The first mouse died after 6 h. Thereafter, 2, 4, 2, and 1 mice died after 12, 24, 36, and 48 h, respectively. No other mice died between 48 and 96 h, and the surviving mice recovered as usual. The total mortality rate was 66.7% . The situation in the C-Δwaafstrain group was similar to that of the WT strain group: the first mouse died after 6 h, and the number of mice that died gradually increased within the first 24 h and then decreased. The total mortality of the C-Δwaafstrain group was 60% (Fig. 6, green line).
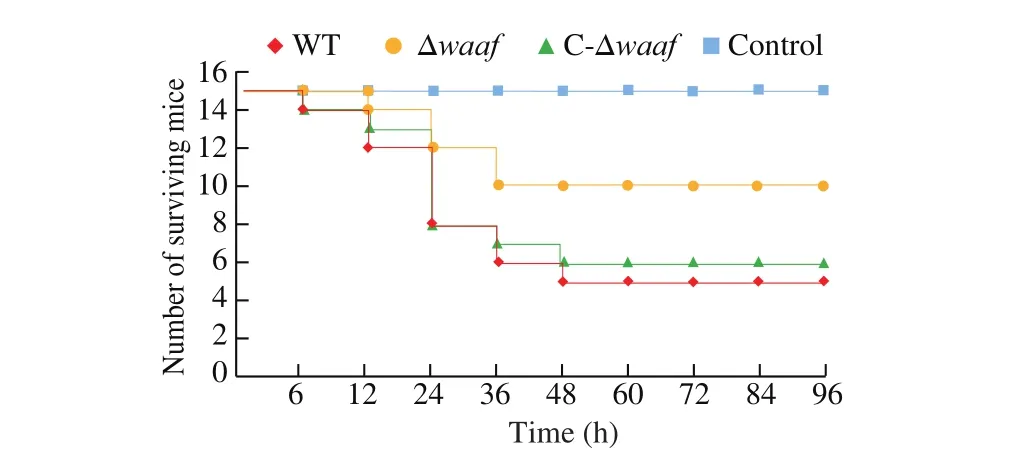
Fig. 6 The quantity of mice surviving in the challenge experiment.
In the Δwaafstrain group, after gavage for 2 h, some mice also showed a state of inactive and reduced diet, but the number and symptoms of mice in the Δwaafstrain group were substantially lower than those in the WT strain group. The first mouse died at 12 h, and then two mice died at 24 and 36 h. No other mouse died within 36–96 h.The total mortality was 33.3% . In the PBS control group, no mice died, and all of them grew normally.
3.5.3 Histopathological observation and intestinal bacterial load
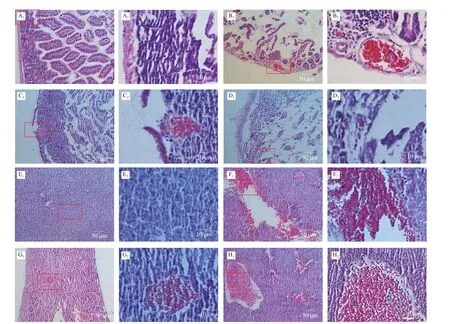
Fig. 7 Pathological sections of small intestine and liver in dead mice. (A–D), Pathological sections of small intestine in dead mice which were given different strain by oral administration. A1, A2 were control (PBS solution); B1, B2 were WT strain; C1, C2 were Δwaaf strain; D1, D2 were C-Δwaaf strain. A2–D2 were partial enlargement of the position indicated with red frame on A1–D1, respectively. (E–H), Pathological sections of liver in dead mice. E1, E2 were control (oral administration PBS solution); F1,F2 were WT strain; G1, G2 were Δwaaf strain; H1, H2 were C-Δwaaf strain. E2–H2 were partial enlargement of the position indicated with red frame on E1–H1, respectively.The wall of small intestine is intact the wall of the small intestine and the villi of the small intestine can be seen clearly.
The cause of death was further investigated by sectioning and observing the tissues small intestines and liver. The small intestine tissues in the control group (gavage with PBS) were regular and closely arranged, and the intestinal wall structure was complete(Figs. 7A1and A2). In the WT strain group, the small intestine tissues were obviously damaged, the intestinal wall became thinner (even broken), the intestinal cells were loose and disordered, the small intestine cells were cracked, and numerous reddish granules appeared in the cytoplasm. Moreover, the inflammatory reaction was obvious(Figs. 7B1and B2). In the Δwaafstrain group, the intestinal tissues of dead mice also appeared to be disrupted, but the extent of damage was lower than that of the WT strain. The intestinal cells were partially destroyed, the intestinal wall was broken, and light red granules (inflammation) (Figs. 7B1and B2) appeared. Nevertheless,some of the intestinal walls were still intact (Figs. 7C1and C2). The sections of the intestines infected with the C-Δwaafstrains were similar to those of the WT strains. Most of the intestinal cells were broken, and the intestinal wall became thinner (Figs. 7D1and D2). The logarithm ofV.parahaemolyticusnumber in the intestinal of mice infected with the WT, Δwaafand C-Δwaafstrains were 6.3 ± 0.3,4.9 ± 0.4 and 5.8 ± 0.3, respectively. These results demonstrated thatV.parahaemolyticuscan adhere the epithelial cells of small intestines and cause disease or death.
The liver cells in the control group were regular and closely arranged (Figs. 7E1and E2). The pathological sections of the liver of mice infected with the WT, Δwaaf, and C-Δwaafstrains were all obviously damaged, and numerous reddish granules appeared in the liver tissues. The extent of damage of the liver infected with the Δwaafstrain was considerably lower than that of the WT and C-Δwaafstrains, and the number and scale of inflammatory reactions substantially decreased (Figs. 7F–H). Thus,V.parahaemolyticuscan enter the host body after invading the small intestines, and the toxin produced byV.parahaemolyticuscan cause inflammation and damage to the liver and other organs.
The results of examination of the tissue sections showed that the damage of the Δwaafstrain to the host was considerably lower than that of the WT strain, consistent with the recorded mortality of mice.These results, combined with those of adhesion assays, the number of bacteria entering the host decreased after the adhesion characteristics of the gene-deficient strains decreased, leading to a decrease in host mortality.
LPS plays an important role in the pathogenesis ofYersinia pseudotuberculosis, in which it has direct toxicity and is an important virulence factor [41]. By contrast, LPS has no direct toxicity inV.parahaemolyticus. The virulence factors ofV.parahaemolyticusinclude TDH, urease, and the T3SS system, among which TDH is the main pathogenic factor [42]. Our study revealed that thewaafgene is vital to LPS synthesis inV.parahaemolyticusand leads to the loss of the O antigen. Deletion of thewaafgene inV.parahaemolyticusresulted in remarkable decreases in the adhesion of pathogenic bacteria to the host and weakened the pathogenicity of this bacterium.InE. coliandHaemophilus parasuis, the deletion of thewaafgene can shorten the side chain of the O antigen polysaccharide, which in turn can weaken the adhesion and invasiveness of these bacteria and then reduce their virulence [43,44]. However, inY. pseudotuberculosis, the deletion of the O antigen synthesis gene can also cause the shortening of the O antigen polysaccharide chain, but it will enhance the virulence of this bacterium [41]. We speculate that the O antigen itself is toxic, and the shortened O antigen polysaccharide chain is more exposed to the core of the O antigen, thereby increasing its toxicity.When the O antigen synthesis gene was knocked out, the toxicity change was closely related to the toxicity of the O antigen itself.
4. Conclusion
Thewaafgene deletion strain was successfully constructed via chitin-mediated genetic transformation. Results showed that thewaafgene can control the serotype ofV.parahaemolyticus. The absence of thewaafgene in the LPS synthesis gene cluster can cause the disappearance of the serotype of O and K, affect the changes in polysaccharides in the outer membrane of the cell, decrease the adhesion of the bacteria, and weaken the pathogenicity ofV.parahaemolyticus. This study is helpful to clarify the pathogenic mechanism ofV.parahaemolyticus.
Funding
This work was supported by grants from the Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX0685), the Science and Technology Research Program of Chongqing Municipal Education Commission (KJQN202001231), the National Natural Science Foundation of China (NSFC) (No. 31201372) and Science and Technology Project of Wanzhou in 2020.
Conflict of interest
The authors declared that they have no conflicts of interest to this work.
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version, at http://doi.org/10.1016/j.fshw.2021.11.026.
- 食品科学与人类健康(英文)的其它文章
- Potential application of proteolysis targeting chimera (PROTAC) modification technology in natural products for their targeted protein degradation
- Effects of vegetarian diet-associated nutrients on gut microbiota and intestinal physiology
- Recent progress in preventive effect of collagen peptides on photoaging skin and action mechanism
- Perspectives on diacylglycerol-induced improvement of insulin sensitivity in type 2 diabetes
- Effect of bacteriocin-producing Pediococcus acidilactici strains on the immune system and intestinal flora of normal mice
- A new Lactobacillus gasseri strain HMV18 inhibits the growth of pathogenic bacteria

