MOLECULAR PHYLOGENETIC EVIDENCES ON MOBILIDA BASED ON GENETIC DISTANCE AND GC CONTENT OF 18S rDNA USING BROAD TAXON SAMPLING
TANG Fa-Hui and ZHAO Yuan-Jun
(Chongqing Key Laboratory of Animal Biology, Chongqing Normal University, Chongqing 401331, China)
MOLECULAR PHYLOGENETIC EVIDENCES ON MOBILIDA BASED ON GENETIC DISTANCE AND GC CONTENT OF 18S rDNA USING BROAD TAXON SAMPLING
TANG Fa-Hui and ZHAO Yuan-Jun
(Chongqing Key Laboratory of Animal Biology, Chongqing Normal University, Chongqing 401331, China)
In order to uncover the real phylogeny of Mobilida ciliates, we sequenced six Trichodinidae species from freshwater fish hosts and acquired nine 18S rDNA sequences. A phylogeny of all Mobilida species with known 18S rDNA sequences was estimated by the maximum likelihood and Bayesian analyses. To our best knowledge, their genetic distance inferred from 18S rDNA was analyzed statistically by using SPSS for the first time. The results further confirmed the non-monophyly of the genus Trichodina and validity of the genus Trichodinella. Additionally, the present study provided 1) new diagnostic evidences combining GC content and genetic distance inferred from 18S rDNA data for family-genus or intraspecific identification of mobilid ciliates and 2) suggested that GC content of 18S rDNA could help to distinguish taxa within Mobilida at family or genus level and was closely related to the divergence of mobilid ciliates. The thresholds of genetic distances inferred from 18S rDNA data for different taxa were proposed to be 0.000—0.005 for intraspecific level, 0.005—0.15 for genus-species levels and higher than 0.15 for family level.
Phylogeny; 18S rDNA; Trichodina; Trichodinella; GC content; Genetic distances
Trichodinid is a diverse mobilid group comprising more than 300 described species of microscopic obligate ectoparasites from maricultured and freshwater animals, typically fishes, molluscs, as well as amphibians[1—12]. According to the classification system of Lynn (2008), the trichodinids have been historically assigned to two major groups of ciliates - now called the Family Trichodinidae and Family Urceolariidae, and the family Trichodinidae is the most important and distributes widely in the Mobilida[13]. All species of this family are characterized by possessing one adhesive disc with some denticles. So far, most studies have still concentrated on the morphological taxonomy and the geographic distribution,whereas, only the morphological studies are not enough to reveal the phlylogenic relationships for the Mobilida[14—20], so more and more valuable evidences, especially the molecular data, are urgently needed to uncover their natural phylogenic position. Up to date, because of lacking enough molecular data in this field, studies on their molecular phylogeny are relatively rare compared with the other ciliates and some important phylogenetic issues have not been resolved such as the natural phylogeny of Trichdodina,the validity of Trichodinella and criteria of familygenus identification within the mobilid ciliates[21—24]. By using broad taxon sampling and supplying new sequences of trichodinids, we analyzed the GC content,genetic distance and phylogenetic tree and presented a further step to disentangle the real phylogenetic position of family Trichodinidae.
1 Materials and methods
1.1 Collection, isolation and morphological identification
The primary sampling sites were along JialingRiver in Chongqing, China (29°5′ N, 106°5′ E). Specimens of host fishes, including Cyprinus carpio,Carassius auratus, Siniperca chuatsi, and Misgurnus anguillicaudatus were collected during February to November, 2007, August, 2008 and March to October, 2009 (Fig. 1). Host fish were examined for trichodinids under the binocular dissecting microscope (NIKON SMZ1500) at×400. Smears of the skin or gill infected by trichodinids were prepared and impregnated by using the dry silver method[25]. Their nuclear apparatus was revealed by using the methyl green-pyronin stain[26]. The impregnated specimens were observated, counted and measured by using a compound microscope (LEICA DM6000B) at 1000× magnification. Systematical and terminological analyses were performed as previously reported[20,27].
Brief introduction of author: Tang Fa-Hui (1979—), Female, Chongqing, Doctor; mainly engaged in fish parasitology;
E-mail: trichodina@126.com
Correspoding author: Zhao Yuan-Jun, E-mail: zhaoyuanjuncqnu@126.com
1.2 DNA isolation, amplification, cloning, and sequencing
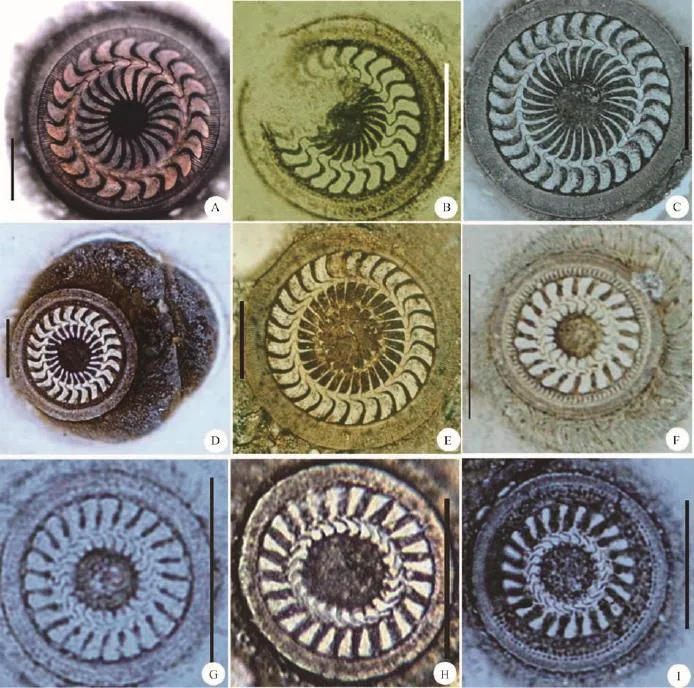
Fig. 1 Morphology of adhesive disc of nine sequenced trichodinids in the present workA. Trichodina paraheterodentata; B. Trichodina pseudominuta; C. Trichodina mutabilis; D. Trichodina modesta; E. Trichodina uniforma;F-K. Four populations of Trichodinella epizootica: F. Trichodinella epizootica (Y); G. Trichodinella epizootica (H); H. Trichodinella epizootica (D); I. Trichodinella epizootica (C). Scale bars: 20 µm
DNA was extracted from each sample of trichodinid species. In brief, after harvested, washed several times in a PCR tube with distilled water and centrifuged at 6000—7500 g, DNA was extracted from 4—5 individuals by using REDExtract-NAmpTM Tissue PCR Kit (Sigma, St. Louis, USA).18S rDNA was amplified by PCR by using the universal eukaryotic primer pair (forward primer 5′-AAC CTG GTT GAT CCT GCC AGT-3′ and reverse primer 5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′) at the following conditions[28]: five cycles of denaturation for 1min at 94℃, primer annealing for 2min at 56℃, and extension for 2min at 72℃, followed by 35 cycles in the same manner, but with the annealing temperature increased to 62℃, and a final extension at 72℃ for 10min. In the case of poor yield by using the universal eukaryotic primer pair, the primer pair MX5-MX3 (forward primer 5′-CTG CGG ACG GCT CAGTAA ATC AGT-3′ and reverse primer 5′-CCA GGA CAT CTT AGG GCA TCA CAGA-3′) was used at the following conditions[29]: 5min initial denaturation at 94℃ followed by 35 cycles of 1min at 94℃, 1min at 56℃, and 2min at 72℃, and a final extention at 72℃ for 10min. The Taq ExDNA polymerase (TaKaRa, Otsu, Japan) was seletcted as the polymerase in all the PCR reaction. Products were purified by using a Gel Extraction Kit,quantified with a NanoDrop ND-1000 spectrophotometer, cloned into pMD19-T vector, and the selected clones were sequenced in an ABI Prism 377 DNA Sequencer (Applied Biosystems Inc., Foster City, Californi).
1.3 Phylogenetic analyses
The nucleotide sequences used for the present analyses are available from GenBank databases (for accession numbers see Tab. 1). A total of 37 complete or partial 18S rDNA sequences, comprising nine newly obtained sequences in this study, were used to construct the phylogenetic trees. The choanoflagellate Proterospongia choanojuncta was selected as the outgroup taxon. All the sequences were assembled using BioEdit 5.0.6, aligned using Clustal X 1.81 and further modified manually using BioEdit[30,31]. Maximum Likelihood (ML) and Bayesian Inference analyses were employed for tree construction. The ML tree was constructed in PAUP*4.0b10[32]. Bootstrap confidence values were calculated with a heuristic search using simple sequence addition and 100 replicates. Bayesian analyses were conducted in MrBayes 3.1.2 under a GTR model with 106generations and tree sampling every 100 generations, and posterior probability distribution was generated using Markov chain Monte Carlo (MCMC) methods[33]. In addition,GC content and genetic distances with Kimura 2-parameter model (Tab. 2) were calculated with MEGA 4.0[34]. In order to explore the family-genus or genus-species relationships among those trichodinids,experimental data of genetic distances were first disposed by software SPSS 16.0 and then analyzed using NPar-Tests in the present work[35]. Fifteen species were selected as the genus Trichodina representatives to test the inter-genus relationship, and thirty groups of genetic distances were used to analyze the relationship of the three genera, viz. Urceolaria,Trichodina and Trichodinella. Moreover, in order to learn about the difference degree of different taxa,NPar-Tests of inter-species or intra-species (interpopulation) relationships were also carried out, and the results of the statistical analysis were listed in Tab. 3—4.
2 Results
2.1 Samples collection information of present sequenced trichodinds (Fig. 1)
Class Oligohymenophora de Puytorac, et al.,1974
Subclass Peritrichia Stein, 1859 Order Mobilida Kahl, 1933 Family Trichodinidae Claus, 1874 Genus Trichodina Ehrenberg, 1838
2.1.1 Trichodina paraheterodentata Tang & Zhao,2013 (GU906244.1)
Host and location: Siniperca chuatsi, skin
Prevalence: Out of 28 fishes examined, five were infected (18%)
Locality: Shapingba, Chongqing (29°59′ N,106°46′ E), China
Date of sampling: August, 2008
2.1.2 Trichodina pseudominuta Tang & Zhao, 2013(HQ407385.1)
Host and location: Carassius auratus, gills
Prevalence: Out of 22 fishes examined, four were infected (18.2%)
Locality: Dazu District, Chongqing (29°57′ N,105°77′ E), China
Date of sampling: October, 2007
2.1.3 Trichodina modesta Lom, 1970 (GU906245.1)
Host and location: Misgurnus anguillicaudatus,gills
Prevalence: Out of 46 fishes examined, twentyone were infected (45.7%)
Locality: Shapingba, Chongqing (29°59′ N,106°46′ E), China
Date of sampling: November, 2007
2.1.4 Trichodina mutabilis Kazubski & Migala, 1968(HQ407384.1)
Host and location: Cyprinus carpio, gills
Prevalence: Out of 21 fishes examined, three were infected (14.3%)

Tab. 1 List of 18S rDNA sequences used in the present work
Locality: Kaixian District, Chongqing (31°18′ N,108°37′ E), China
Date of sampling: March, 2009
2.1.5 Trichodina uniforma Van AS & Basson, 1989(HQ407383.1)
Host and location: Carassius auratus, gills
Prevalence: Out of 65 fishes examined, eighteen were infected (27.7%)
Locality: Dazu District, Chongqing (29°57′ N,105°77′ E), China
Date of sampling: August, 2008 Family Trichodinidae Claus, 1874 Genus Trichodinella Sramek-Husek, 1953 Species Trichodinella epizootica Sramek-Husek, 1953
2.1.6 Population one: Trichodinella epizootica (Y)(HQ407388.1)
Host and location: Misgurnus anguillicaudatus,gills
Prevalence: Out of 46 fishes examined, twentyeight were infected (60.9%)
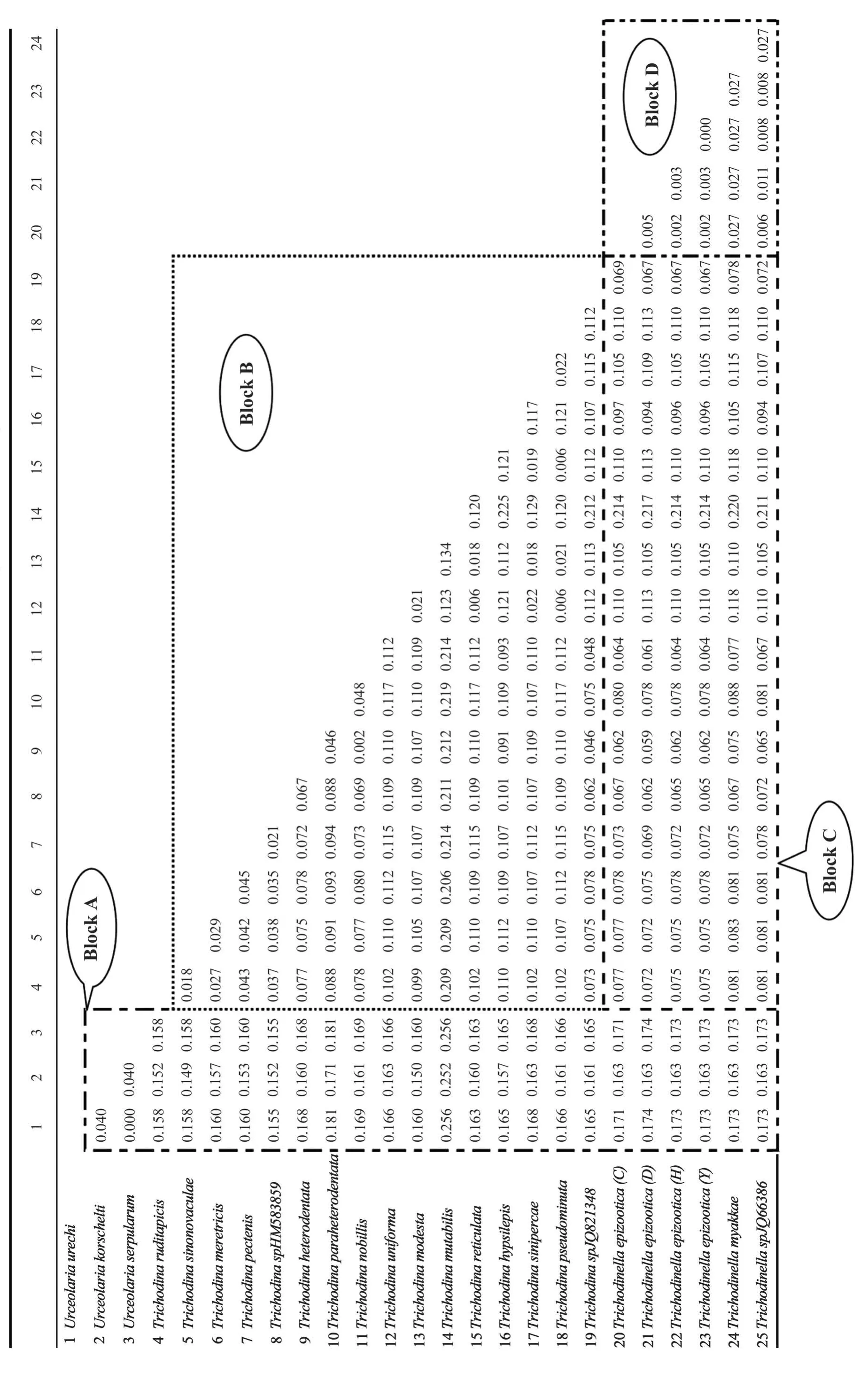
Tab. 2 Test results of SPSS statistics based on the genetic distance data
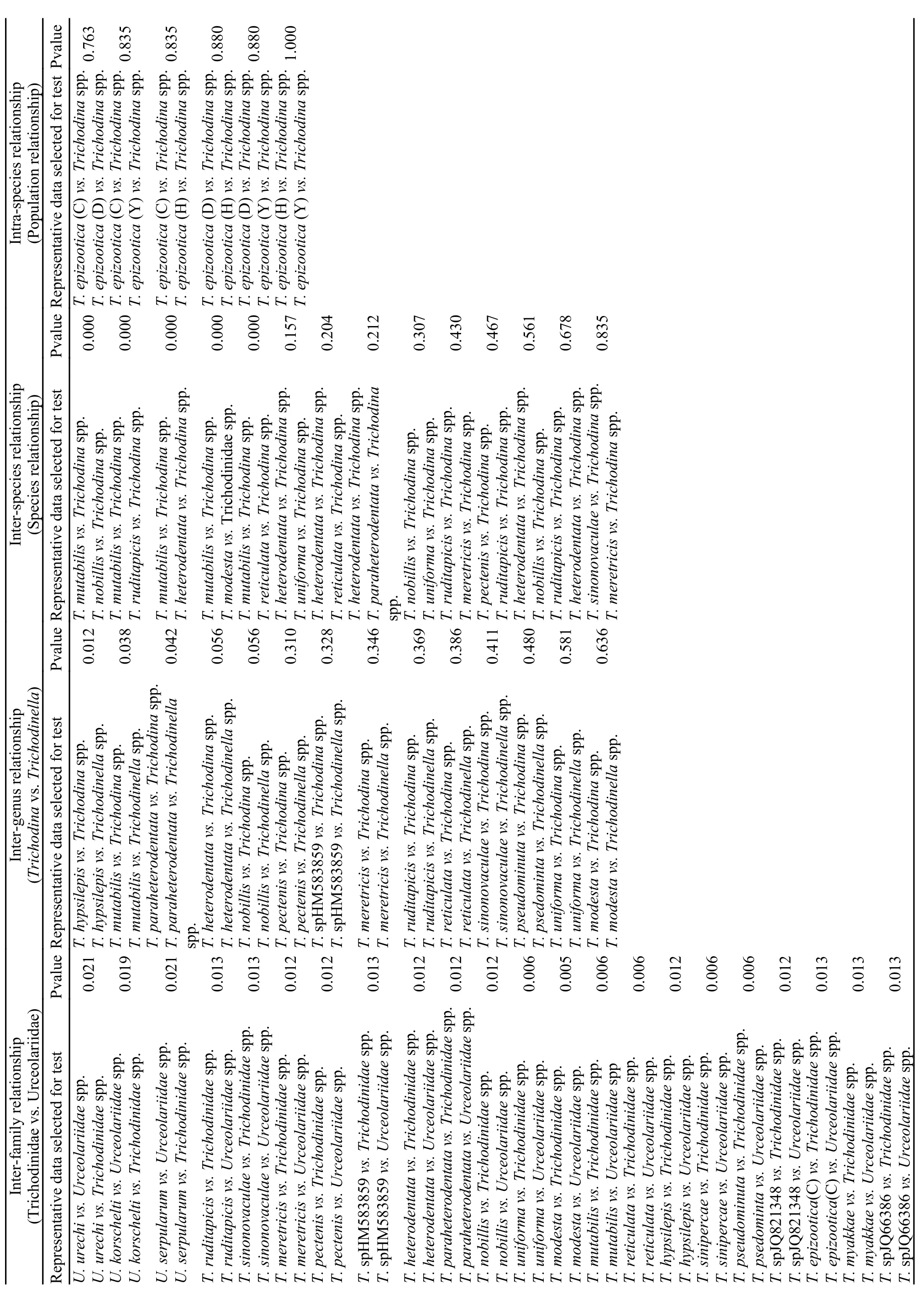
Tab. 3 Test results of SPSS statistics based on the genetic distance data

Tab. 4 Nucleotide composition and GC contents of 18s rDNA se-quences of Mobilida specise
Locality: Yanggongqiao, Shapingba, Chongqing(29°56′ N, 106°44′ E), China
Date of sampling: November, 2007
2.1.7 Population two: Trichodinella epizootica (H)(GU906246.1)
Host and location: Carassius auratus, gills
Prevalence: Out of 38 fishes examined, 22 were infected (57.9%)
Locality: Hulongba, Chongqing (29°71′ N,106°41′ E), China
Date of sampling: June, 2007
2.1.8 Population three: Trichodinella epizootica (D)(HQ407387.1)
Host and location: Cyprinus carpio, gills
Prevalence: Out of 18 fishes examined, four were infected (22.2%)
Locality: Dianjiang District, Chongqing (30°28′N, 107°48′ E), China
Date of sampling: February, 2007
2.1.9 Population four: Trichodinella epizootica (C)(HQ407386.1)
Host and location: Carassius auratus, gills
Prevalence: Out of 89 fishes examined, thirtyfour were infected (38.2%)
Locality: Chenjiaqiao, Chongqing (29°64′ N,106°37′ E), China
Date of sampling: October, 2009
2.2 Genetic distances within collections and SPSS analyses
As shown in Tab. 2, the genetic distances among all the mobilid species was ranged from the minimum of 0.000 between Urceolaria serpularum and Urceolaria urechi to the maximum of 0.256 between Urceolaria serpularum and Trichodina mutabilis as well as between Urceolaria urechi and Trichodina mutabilis. The genetic distances were 0.150—0.256 between the Family Urceolariidae (eg. Urceolaria species) and Trichodinidae (eg.Trichodina or Trichodinella species) (Tab. 2, Block A), 0.002—0.225 among species of the genus Trichodina (Tab. 2,Block B), 0.059—0.220 among different species between genera Trichodina and Trichodinella (Tab. 2,Blcok C) and 0.000—0.027 among species(0.006—0.027) or populations (0.000—0.005) of the genus Trichodinella (Tab. 2, Block D).
The data of genetic distances were further used to test the relationship of the different taxa in mobilida with SPSS statistics (Tab. 2). The statistical results showed that all P-values in SPSS analysis were less than 0.05 (ranging from 0.005—0.021) between family Trichodinidae and Urceolariidae. However,most P-values between genus Trichodina and Trichodinella were greater than 0.05, ranging from 0.012 to 0.636, and nearly all P-values of inter-species or intra-species (population) were higher than 0.05, ranging from 0.157—0.835 except the P values,which were all 0.000, between Trichodina mutabilis and other species. In addition, the P-values among populations of Trichodinella epizootica are much higher, most of which were close to or more than 0.800, even reaching 1.000 (Tab. 3).
2.3 GC content analyses
The GC contents of 18S rDNA for the 25 mobiline species used in the phylogenetic analyses are listed in Tab. 4. Among genus Trichodina of the family Trichodinidae, GC content was the highest in Trichodina nobillis and the lowest in Trichodina modesta, and its overall variation was in a wide range between 46.43%—50.82%. However, genus Trichodinella possesses the relatively higher GC content(ranging from 50.26% to 50.58%) when comparedwith other groups. The GC content in the family Urceolariidae studied here was clearly lower varying between 43.70%—44.10%.
The species with GC content higher than 50% mainly include all the Trichodinella species, and three Trichodina species, viz. Trichodina nobillis, Trichodina paraheterodentata and Trichodina heterodentata, all of which are freshwater trichodinids. The species with GC content between 46% and 50% include eight freshwater and five marine Trichodina species. The species with GC content lower than 46% were consisted of three Urceolariidae species,Urceolaria urechi, Urceolaria serpularum and Urceolaria korschelti.
2.4 Phylogenetic analyses
The Peritrichia molecular data were used for phylogenetic trees of Maximum likelihood and Bayesian inference. The phylogenetic analysis results produced a well-resolved and strongly supported single optimal cladogram. Analyses using ML and BI methods reached almost same topology of phylogenetic tree and identical higher-level relationships along the backbone (Fig. 2, 3).
According to the phylogenetic tree’s topology out-grouped with Proterospongia choanojuncta,Clade I (Mobilida) and Clade II (Sessilida) were fully recovered in the present study (Fig. 3). In the Mobilida clade, all the Urceolariidae species (Urceolaria species) and Trichodinidae species (Trichodina and Trichodinella species) grouped together respectively. The Urceolaria clade branched the most basal position to the out-group in the cladogram (Fig. 3), and within the genera Urceolaria, Urceolaria urechi and Urceolaria serpularum first clustered together, then clustered with Urceolaria korschelti.
Additionally, all Trichodinella epizootica populations grouped together with bootstrap support of 95% in ML and a posterior probability of 1.00 in the Bayesian analysis. The population C (HQ407386) and D (HQ407387) had nearly identical sequence despite being collected from different hosts at sites slightly more than 130 km apart in Chongqing area. Trichodinella sp. (JQ663869) was separated from Trichodinella epizootica and preceded by Trichodinella myakkae (AY102176) with bootstrap support of 77% and 100% in ML and posterior probability of 0.92 and 1.00 in the Bayesian analysis. All Trichodinella isolates consistently branched off the top of the Trichodina cluster comprising Trichodina heterodenta,Trichodina nobillis, Trichodina paraheterodentata and Trichodina sp. JQ821348. Remarkably, all the marine Trichodina species, Trichodina ruditapicis,Trichodina sinonovaculae, Trichodina meretricis, Trichodina pectenis and Trichodina sp. HM583859 always formed one independent cluster although they were also included in other freshwater clusters (Fig. 2,3).
3 Discussion
3.1 Confirmation of the non-monophyly of the genus Trichodina
The genus Trichodina is the largest genus in the family Trichodinidae and its real phylogenetic position has been a research focus in the field. Although some research work tried to explore the relationships between the blade morphological characters and the systematic differentiation[22], and some studies even ambiguously considered Trichodina as a monophyly[24],there is no exact or explicit statement about the real phylogeny of genus Trichdodina up to now.
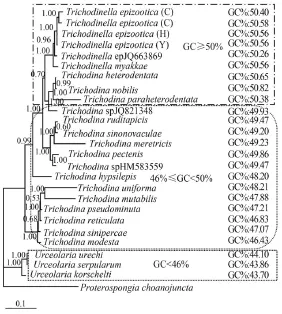
Fig. 2 The Bayesian phylogenetic tree of 25 mobilid species with GC contents labeled
In the present molecular analyses, all the analytic results including GC content, genetic distance and phylogenetic tree further corroborate that the large genus Trichodina is non-monophyletic based on the following reasons. 1) The genus Trichodina possesses the widest range of GC content(46.43%—50.82%) when compared with the genus Trichodinella and genus Urceolaria (Tab. 4, Fig. 2). The GC content of the genus Trichodinella is higher than 50%, ranging from 50.26%—50.58%, while that of the genus Urceolaria is lower than 46%, ranging from 43.7%—44.1%. According to the relationship of GC content influencing the species differentiation in this group[22], the genus Trichodina with GC content variety can not be monophyletic in the family Trichodinidae. 2) From the visual angle of genetic distances analysis, the genetic distances between dif-ferent Trichodina species varied widely, ranging from 0.002 to 0.219, whereas both genetic distances of two other genera Trichodinella (ranging from 0.006—0.027) and Urceolaria (ranging from 0.000—0.040) varied narrowly, which indicated that the speciation processes for genus Trichodina were not synchronous. Furthermore, statistical results indicated that the significance of difference in genetic distance between Trichodina and Trichodinella was uncertain (P=0.01—0.70). In addition, although the difference in genetic distance of Trichodina hypsilepis and Trichodina mutabilis to Trichodinella was significant (P=0.012 vs. P=0.038), the differences in genetic distances of most Trichodina species (Tab. 3),such as Trichodina modesta, Trichodina unifroma to Trichodinella were not significant (P=0.056—0.636),indicating that the position of monophyly for genus Trichodina was uncertain. 3) In the ML/BI phylogenetic trees, although Trichodinidae formed one clade,all the Trichodina species were scattered and separated into several diverged clades with well or strong support in the phylogenetic trees. They did not cluster as one monophyletic clade and were mixed with Trichodinella species, which further supported the previous results and proved that the genus Trichodina was not monophyletic.
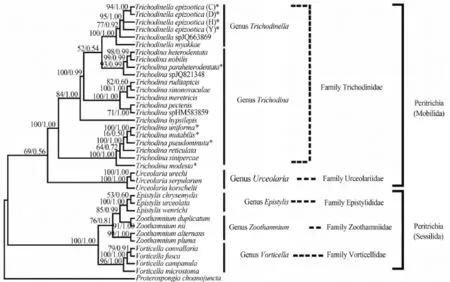
Fig. 3 Phylogenetic tree based on small subunit rDNA sequences by Maximum Likelihood (ML) and Bayesian Inference (BI) with the model of ‘‘GTR+I+G’’
3.2 Validity re-evaluation of genus Trichodinella
In the past several years, many efforts have been made to explore the phylogeny of family Trichodinidae; however, relationships within this group remain unclear. Among the 11 genera of the family Trichodinidae[20], the genus Trichodinella is nearly the smallest and most specialized group because of the short and curved denticle in the adhesive disc,which obviously differs from the adhesive disc characters of the genus Trichodina. Additionally,Trichodinella was morphologically distinguished from other genera by the difference of the adoral spiral (180°—270° in Trichodinella vs. 360—540° in Tricodina)[36]. However, recent molecular data have challenged the validity of the genus Trichodinella[22],because in all the molecular phylogenetic trees,Trichodinella always nest within the genus Trichodina with full or very high node support, rather than form into one independent clade[22,24]. It is hard for previous studies to evaluate the validity of genus Trichodinella and uncover its real phylogenetic position mainly because of the low Trichodinella sampling.
Now by broad taxon sampling of Trichodinella and adding much more DNA sequences, the phylogenetic study on Trichodinella was re-proceeded. The present analyses support the establishment of the genus Trichodinella for the following reasons. 1) In the phylogenetic trees, it appeared that some Trichodina species scattered with Trichodinella ones by adding new sequences; however, Trichodinella still forms a well-supported monophyletic clade although it still nests within Trichodina. 2) In the case of GC content of 18S rDNA, the genus Trichodinella possessing higher GC content, and GC content of all species in this genus was higher than 50% (Tab. 4, Fig. 2), which supports that the Trichodinella as one independent genus is different from Trichodina. 3) The other important aspect supporting the validity of genus Trichodinella could be still the morphological characteristics and the parasitic specificity when compared to genus Trichodina. Trichodinella species possess smaller body size (less than 30 μm vs. more than 30 μm), shorter adoral ciliary spiral turns(180°—270° vs. 360°—540°), more undeveloped or degenerated ray (absent or short and curved vs. long and developed) and is gill specialized parasite (vs. gill, skin, fin et al.).
3.3 New diagnostic evidences for family-genus or intraspecific identification for mobilid ciliates
The studies on GC content or base composition in different organisms have been reported before[37—40]. The present study combined GC content and genetic distance inferred from 18S rDNA data to analyze different family or genus of mobilid ciliates. It is important to note that consistent with previous studies, GC content of 18S rDNA was different between families Urceolariidae and Trichodinidae: which was less than 45% in the former, but significantly higher than 45% in the latter. In addition, in family Trichodinidae, GC content was more uniform in the genus Trichodinella,almost all exceeding 50%, than that of genus Trichodina, which varied significantly among species (Fig. 2). By this token, GC content of 18S rDNA could be used to aid in distinguishing family-genus within Peritrichia. Moreover, it was revealed again that GC content influences the structure of phylogenetic tree,which also further corroborated the previous viewpoint that GC content was closely related to the divergence of mobilid ciliates[22].
The analysis of genetic distance for phylogeny in different organisms has been previously reported[41]. According to the current analysis results (Tab. 2), the genetic distance range between families Urceolariidae and Trichodinidae is 0.150—0.256, whereas the
[1]Lom J, Dykova I. Protozoan parasites of fishes [M]. Amsterdam: lsevier Science Publishers B. V. 1992, 1—315
[2]Lom J. Trichodinid ciliates (Peritrichida: Urceolariidae) from some marine fishes [J]. Folia Parasitologica, 1970, 17: 113—125
[3]Van As J G., Basson L. A further contribution to the taxonomy of Trichodinidae (Ciliophora: Peritrichia) and a regenetic distance is 0.000—0.040 within Urceolariidae and 0.002—0.225 within Trichodinidae. SPSS statistic analyses (Tab. 3) indicated that there is a significant difference in genetic distance between families Urceolariidae and Trichodinidae(P=0.005—0.021), but not between Trichodina and Tricho- dinella (P=0.006—0.636). Therefore, genetic distance should be considered as an important identification basis for inter-family classification within mobilid ciliates, but not for inter-genus classification within family Trichodinidae. Besides, genetic distance could be used to help diagnosing those intraspecific or close interspecific groups. To find the optimal thresholds for intraspecific genetic distances for different groups, given the exceptionally maximum genetic distance ascribed to Trichodina mutabilis,three thresholds of genetic distances inferred from 18S rDNA data for different taxon were concluded based on the present research. According to the analysis, the intraspecific genetic distances should be 0.000—0.005, and genus-species genetic distances should be 0.005—0.15. When genetic distance is greater than 0.15, the diffe- rence reaches the family level. Additionally, the re- sults of genetic distance analyses challenged the morphological taxonomy of some species. For example, the genetic distance between T. nobilis and T. heterodenta was only 0.002,which fell into the intraspeci- fic coverage on the basis of the above conclusion. So we further blasted and compared their sequence alignment and found that their 18S rDNA genes had same length (1698 nt)and 99% similarity with only three base difference. We speculate that this is due to either morphological misidentification or sequencing error. It was unexpected that the genetic distance of Urceolaria urechi and Urceolaria serpularum was 0.000. Combined their nucleotide composition of 18S rDNA sequences with 100% sequence similarity, these two species,Urceolaria urechi and Urceolaria serpularum, were proved the same species. In summary, the levels of detected variation are probably the combination of intraspecific variation and experimental error, including the occasional contamination of SSU rDNA sequences and misidentification.view of the taxonomic status of some fish ectoparasitic trichodinids [J]. Systematic Parasitology, 1989, 14(3): 157—179
[4]Van As J G, Basson L. Trichodinid ectoparasites (Ciliophora: Peritrichida) of freshwater fishes of the Zambesi river System, with a reappraisal of host specificity [J]. Systematic Parasitology, 1992, 22(2): 81—109
[5]Song W B. Pathogenic Protozoa in Mariculture [M]. Beijing: Science Press. 2003, 429—483 [宋微波. 海水养殖中的危害性原生动物. 北京: 科学出版社. 2003, 429—483]
[6]Zhao Y J, Tang F H. Trichodinid ectoparasites from the freshwater fish Misgurnus anguillicaudatus (Cantor) and Mollusc Anodonta woodiana (Lea) of Chongqing in China,with descriptions of two new species of Trichodina Ehrenberg, 1838 [J]. Systematic Parasitology, 2007, 67(1): 65—72
[7]Tang F H, Zhao Y J, Chen H. Trichodinid ectoparasites from golden carp, with a description of Trichodina paranigra sp. nov. [J]. Acta Hydrobiologica Sinica, 2005, 29(1): 74—80[唐发辉, 赵元莙, 陈辉. 鲫寄生车轮虫一新种的描述. 水生生物学报, 2005, 29(1): 74—80]
[8]Tang F H, Zhao Y J, Tang A K. Presence of ectoparasitic trichodinids (Ciliophora, Oligohymenophorea, Peritrichida)on the gills of cultured freshwater fish, Carassius auratus in Chongqing, China, with the the description of a new species of the genus Trichodina [J]. Acta Zootaxonomica Sinica,2005, 30(1): 35—40
[9]Tang F H, Zhao Y J, Tao Y F. Trichodinids (Ciliophora: Peritrichida) parasitic on gills of freshwater fishes, Carassius auratus and Aristichthys nobilis from China, with the description of Trichodina subtilihamata sp. nov. [J]. Zootaxa,2007, 1582: 39—48
[10]Tang F H, Zhao Y J. Study of trichodinids (Protozoa, Ciliophora) parasitic on gills of freshwater fishes from Chongqing, China, and identification of a new species,Trichodina cyprinocola sp. nov.[J] African Journal of Microbiology Research, 2011, 5(26): 5523—5527
[11]Tang F H, Zhao Y J, Liu C N. First records of three Tripartiella species (Ciliophora, Oligohymenophora, Peritrichida) from freshwater fishes along Yangtze River in China[J]. Zootaxa, 2013, 3681: 169—174
[12]Tang F H, Zhao Y J, Liu C N. Two trichodinids of Paratrichodina Lom, 1963 (Ciliophora, Peritrichida, Trichodinidae) infecting gills of Ietalurus punetaus from Chongqing,China [J]. African Journal of Microbiology Research, 2012,6(9): 2145—2149
[13]Lynn D H. The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature (3rd) [M]. Springer,Dordrecht, 2008, 428—435
[14]Lom J. Observations on trichodinid ciliates from freshwater fishes [J]. Archiv für Protistenkunde, 1970, 112: 153—177
[15]Lom J, Hoffman G L. Geographic distribution of some species of trichodinids (Ciliata: Peritricha) parasitic on fishes[J]. Journal of Protozoology, 1964, 50: 30—35
[16]Lom J, Laird M. Parasitic protozoa from marine and euryhaline fish of Newfoundland and New Brunswick. I. Peritrichous ciliates [J]. Canadian Journal of Zoology, 1969, 47: 1367—1380
[17]Lom J. Trichodinid ciliates from fishes of the Rumanian Black Sea Coast [J]. Parasitology, 1962, 52: 49—61
[18]Zhao Y J, Tang F H. Taxonomic study on trichodinids (Protozoa, Ciliophora) infecting on gills of freshwater fishes,Cyprinus carpio and Mylopharyngodon piceus from China,with the description of Trichodina regularis sp. nov. [J]. European Journal of Scientific Research, 2011, 58(2): 231—237
[19]Tang F H, Zhao Y J. Taxonomic study on trichodinids parasitic on gills of freshwater fish, Carassius auratus from Chongqing, China, with the description of Trichodina brevicirra sp. nov. [J]. Acta Hydrobiologica Sinica, 2010, 34(5): 1004—1011
[20]Tang F H, Zhao Y J. Record of three new Trichodina species (Protozoa, Ciliophora) parasitic on gills of freshwater fishes from Chongqing, China [J]. African Journal of Microbiology Research, 2013, 7(14): 1226—1232
[21]Utz L R P, Eizirik E. Molecular phylogenetics of subclass Peritrichia (Ciliophora: Oligohymenophorea) based on expanded analyses of 18S rRNA sequences [J]. Journal of Eukaryotic Microbiology, 2007, 54(3): 303—305
[22]Tang F H, Zhao Y J, Warren A. Phylogenetic analyses of trichodinids (Ciliophora, Oligohymenophora) inferred from 18S rRNA gene sequence data [J]. Current Microbiology,2013, 66(3): 306—313
[23]Gong Y, Yu Y, Li X, et al. Reevaluation of the phylogenetic relationship between mobilid and sessilid peritrichs (Ciliophora, Oligohymenophorea) based on small subunit rRNA genes sequences [J]. Journal of Eukaryotic Microbiology,2006, 53(5): 397—403
[24]Zhan Z F, Xu K D, Dunthorn M. Evaluating molecular support for and against the monophyly of the Peritrichia and phylogenetic relationships within the Mobilida (Ciliophora,Oligohymenophorea) [J]. Zoologica Scripta, 2013, 42(2): 213—226
[25]Klein B M. The dry silver method and its proper use [J]. Journal of Protozoology, 1958, 5: 99—103
[26]Foissner W. Basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa [J]. European Journal of Protistology, 1991, 27: 313—330
[27]Corliss J O. The Ciliated Protozoa (2nd ed) [M]. Oxford: Pergamon Press, Elmsford NY. 1979, 1—128
[28]Medlin L, Elwood H, Sticke S, et al. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions [J]. Gene, 1988, 71(2): 491—499
[29]Andree K, SzéKely C, Molná K, et al. Relationships among members of the genus Myxobolus (Myxozoa: Bilvalvidae)based on small subunit ribosomal DNA sequences [J]. Journal of Protozoology, 1999, 85(1): 68—74
[30]Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT [J]. Nucleic Acids Symposium Series, 1999, 41: 95—98
[31]Thompson J D, Gibson T J, Plewniak F, et al. The Clustal X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools [J]. Nucleic Acids Research, 1997, 25(24): 4876—4882
[32]Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods), version 4.0b10 [M]. Sinauer Associates, Sunderland, Massachusetts. 2003
[33]Ronquist F, Huelsenbeck J. MRBAYES 3: Bayesian phylogenetic inference under mixed models [J]. Bioinformatics,2003, 19(12): 1572—1574
[34]Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods [J]. Molecular Biology and Evolution, 2011, 28(10): 2731—2739
[35]Morgan G A, Leech N L, Gloeckner G W, et al. SPSS for Introductory Statistics: Use and Interpretation [M]. Mahwah,NJ: Lawrence Erlbaum Associates. 2004
[36]Basson L, Van As J G. Differential diagnosis of the genera in the family Trichodinidae (Ciliophora: Peritrichida) with the description of a new genus ectoparasitic on freshwater fish from southern Africa [J]. Systematic Parasitology, 1989, 13: 153—160
[37]Smit S, Widmann J, Knight R. Evolutionary rates vary among rRNA structural elements [J]. Nucleic Acids Research, 2007, 35(10): 3339—3354
[38]Cao J, Wu X, Jin Y. Lower GC-content in editing exons: implications for regulation by molecular characteristics maintained by selection [J]. Gene, 2008, 421(1—2):14—19
[39]Du H, Hu H, Meng Y, et al. The correlation coefficient of GC content of the genome-wide genes is positively correlated with animal evolutionary relationships [J]. FEBS Letters,2010, 584(18): 3990—3994
[40]Mooers A, Holmes E C. The evolution of base composition and phylogeneticinference [J]. Trends in Ecology and Evolution, 2000, 15(9): 365—369
[41]Zhao Y J, Li N N, Tang F H, et al. Remarks on the validity of Myxobolus ampullicapsulatus and Myxobolus honghuensis (Myxozoa: Myxosporea) based on SSU rDNA sequences [J]. Parasitology Research, 2013, 112: 3817—3823
10.7541/2016.48
基于18S rDNA遗传距离与GC含量对游走类纤毛虫的分子系统学研究
唐发辉 赵元莙
(重庆师范大学, 重庆师范大学动物生物学重点实验室, 重庆 401331)
为了揭示游走类纤毛虫的系统发生, 对寄生于淡水鱼类的车轮虫科中的6种车轮虫进行了18S rDNA的测序并获得了9个序列。采用了最大似然法(ML)与贝叶斯法(BI)对GenBank中所有游走类纤毛虫的18S rDNA序列进行了系统树的构建, 并首次将SPSS与18S rDNA遗传距离结合分析了游走类纤毛虫的系统发生。研究结果进一步证实了车轮虫属(Trichodina)的非单系发生与小车轮虫属 (Trichodinella) 的有效性。此外, 研究结合18S rDNA 的GC含量与遗传距离分析提出了游走类纤毛虫科属及种间新的鉴定依据: 18S rDNA 的GC含量可用于游走类纤毛虫的科属区分, 且与游走类纤毛虫的分化密切相关; 18S rDNA的遗传距离在游走类纤毛虫的不同阶元中具有一定的阈值范围, 即通常种内遗传距离阈值范围为0.000—0.005, 属种间阈值范围为0.005—0.150, 当遗传距离大于0.150时, 则达到了科间水平。
系统发生; 18S rDNA; 车轮虫属; 小车轮虫属; GC含量; 遗传距离
Q959.116 Document code: A Article ID: 1000-3207(2016)02-0358-12
date: 2015-04-10; Accepted date: 2015-08-24
The National Natural Science Foundation of China (No. 31101637, No. 31172068, No. 31471980); the project of Chongqing Science & Technology Commission (No. CSTC, 2010CA1010; No. cstc2014jcyjA80008); the Science Research Foundation of the Education Committee of Chongqing (No. KJ1400530)

