Effect of GW8 Gene Editing on Appearance Quality of Erect-Panicle Type (dep1) Japonica Rice
Mao Ting, Chen Hongfa, Li Xin, Liu Yan, Zhong Shuncheng, Wang Shiyu, Zhao Yizhou, Zhang Zhan, Ni Shanjun, Huang He, Li Xu, Hu Shikai
Letter
Effect ofGene Editing on Appearance Quality of Erect-Panicle Type ()Rice
Mao Ting1, 2, Chen Hongfa2, Li Xin1, Liu Yan1, Zhong Shuncheng1, Wang Shiyu1, Zhao Yizhou1, Zhang Zhan1, Ni Shanjun1, Huang He1, Li Xu1, Hu Shikai2
(Liaoning Institute of Saline-Alkali and Utilization, Panjin 124010, China; State Key Laboratory of Rice Biology and Breeding / China National Center for Rice Improvement, China National Rice Research Institute, Hangzhou 310006, China)
Most high-yieldingrice varieties in China carry, a multi-effective regulator of plant architecture and grain shape, resulting in erect panicle with short and round grain shape. However, its appearance quality needs to be improved since long-grain rice is favored by the market.is a dominant gene regulating grain shape, and its loss-of-function genotype leads to elongated grains with a better quality in appearance. Whether long-grainrice retaining other high quality traits can be created bygene editing under the genetic background ofremains unsystematically analyzed. In this study, a series ofgene editing near-isogenic lines (NILs) were constructed with different target regions on the promoter region (NIL-gw8), the 1stexon region (NIL-gw8) and the miR156 target region of the 3rd exon (NIL-gw8) under the genetic background of.Grain quality and yield components analyses showed that the grain length to width ratio ofediting offspring increased significantly, and the chalkiness degree decreased obviously, however, owing to the decrease of 1000-grain weight and effective panicle number, the yield also showed a significant downward trend. Among all thegene editing offspring plants, those edited at the miR156 target region showed the largest grain length to width ratio, the lowest chalkiness degree and the best eating quality. These results indicated that the miR156 target region editing of thegene had the best effect on improving the appearance quality of erect-panicle typerice.
Rice is one of the most important staple food crops, feeding more than half of the global population. Grain shape affects both yield and quality, and its genetic improvement and gene mining have gained much attention (Li et al, 2018; Liu et al, 2018).is a positive regulator of grain width (Wang et al, 2012), encoding a squamosa promoter binding protein domain-containing transcription factor, OsSPL16, which regulates grain width by affecting the proliferation of glume cells. High expression ofpromotes cell division and grain filling rate, thereby promoting grain width and yield (Sun et al, 2018). Loss-of-function ofin Basmati rice leads to elongated grains and improved appearance quality. There are two loss-of-function alleles in, including a 10-bp deletion type in the promoter region and a 2-bp deletion in the miR156 region of the 3rd exon (Wang et al, 2012). However, the loss-of-function alleles ofarevery little distribution inrice (Yi et al, 2016; Mao et al, 2021).Conversely, theallele is widely used in high-yieldingrice breeding, resulting in short and round grain shape (Xu et al, 2014). According to previous studies, editing of different grain shape genes is greatly influenced by the genetic background of(Mao et al, 2021; Tao et al, 2023).Therefore, the genetic effect ofshould be considered comprehensively to improve appearance quality ofrice by introducing long-grain shape alleles.
In this study, we designed three target loci in: the promoter region, the 1st exon region and the miR156 target region inthe 3rd exon, in an erect-panicle type (genotype)rice variety Yanfeng 47 (hereinafter referred to as NIL-YF47), and evaluated the application potential of thegene in breeding of long-grainrice by determining the yield and quality of the genetically edited ricematerials with different target loci, which would enrich the theoretical basis for the genetic improvement ofrice quality. First, we sequencedthe target sites of CRISPR-Cas9 edited rice materials.The results indicated that the mutations had been introduced into different target sites of, and all of them were single base insertion types(Fig. S1-A and -B). Except for the promoter mutation, which cannot predict the amino acid changes, the other mutations caused premature termination of amino acid translation at different positions(Fig. S1-A and -C). Also, thegene expression was significantly reduced in the glume cells ofthe editing offspring at 2 d before heading (Fig. S1-D). Further, the basic characteristics of the gene edited materials were examined, including the plant architecture, whole growth duration, plant height and the number of leaves in main stem (Fig. S1-E to -H). The genetic background control, NIL-YF47, had 162 d whole growth period,with a plant height of 99.1 cm and the mean number of leaves in main stem of 15. In all theedited materials, these characteristics remained insignificantly different from NIL-YF47. Subsequently, the NILs of theedited materials were constructed, including NIL-gw8, NIL-gw8and NIL-gw8.
The grain shape was compared among the NILs (Fig. 1-A to -E).For grain length, relative to NIL-YF47(4.88 mm), the longest grains were found in NIL-gw8of 5.33 mm, followed by NIL-gw8and NIL-gw8with the grain lengths of 5.23 and 5.06 mm, respectively. The grain widths of NIL-gw8, NIL-gw8and NIL-gw8were 2.64, 2.31 and 2.28 mm, respectively. The variations in grain length and width led to alterations in the grain length to width ratio, with NIL-gw8showing the highest of 2.33, followed byNIL-gw8and NIL-gw8of 2.19 and1.98, respectively, and all of which were significantly different from that of NIL-YF47(1.78). To explore the factors influencing grain shape in the NILs, we analyzed the changes in glume cell length, width and the key regulatory hormones at 2 d before heading. Compared with NIL-YF47, all theediting offspring showed significantly greater glume length with shorter glume cell width (Fig. 1-F to -H).Moreover, the key regulatory hormones for glume development, indole-3-acetic acid (IAA), gibberellin A3(GA3) and brassinolide (BR) were all significantly lower in theediting offspring(Fig.1-I to -K).
Fig. 1. Comparison of grain shape of near-isogenic lines (NILs) and analysis of relevant influencing factors.
A and B, Performances of grain length (A) and grain width (B) of NILs. Scale bars, 5 mm. C‒E, Comparisons of grain length (C), grain width (D) and grain length to width ratio (E) of NILs. F‒H, Comparisons of glume epidermal cell size (F), cell length (G) and cell width (H) of NILs. Scale bar, 10 μm. I‒K, Comparisons of the levels of indole-3-acetic acid (IAA) (I),gibberellin A3(GA3) (J) and brassinolide (BR) (K) in developing glumes of NILs.Data are Mean ± SD (= 9).
Different lowercase letters above the bars indicate significant differences at< 0.05 determined by the Student- Newman-Keuls test.
After being processed into milled rice, the chalkiness degrees of the NILs were examined (Fig. 2-A and -B). Compared with NIL-YF47(7.06%), the chalkiness degrees of all theediting offspring were significantly lower, with NIL-gw8showing the lowest at 1.50%, followed by NIL-gw8(2.11) and NIL-gw8(3.58%). Numerous studies have shown thatgrain filling characteristics have a strong influence on chalkiness formation (Wang et al, 2008; Li et al, 2014). Therefore, we analyzed the effects of average grain filling rate as well as related hormones and enzymes at the peak grain filling stage (10 d after fertilization) on chalkiness formation. The average grain filling rate was the highest in NIL-YF47, followed by NIL-gw8, NIL-gw8and NIL-gw8, which were consistent with the variations in chalkiness degree (Fig. 2-C). The four starch-filling enzyme activities of adenosine diphosphateglucose (ADPG), granule-bound starch synthase (GBSS), starch branching enzymes (SBE) and soluble starch synthase (SSS) were also in good agreement with the average grain filling rates (Fig. 2-D to -G). Several key hormone levels in the gene editing offspring were also significantly reduced (Fig. 2-H to -L), but the degree of agreement with the grain filling rate was not as good as that of starch-related enzymes. It is worth noting that NIL-gw8, which displayed the lowest chalkiness degree, also showed lower hormone levels with the exception of IAA.
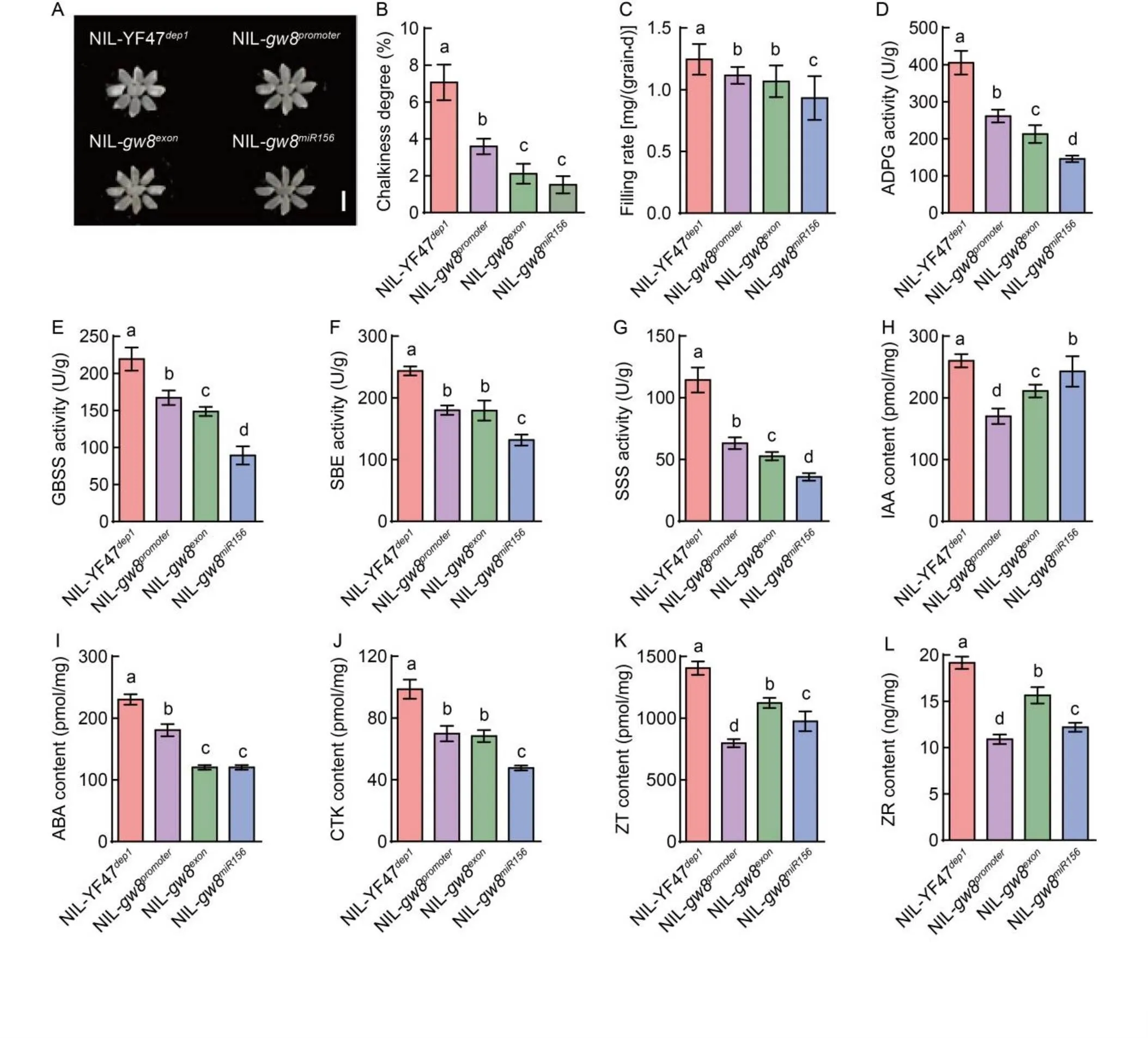
Fig. 2. Comparison of chalkiness degree of near-isogenic lines (NILs) and analysis of releted influencing factors.
A and B, Comparisons of chalkiness degree in endosperms of NILs. Scale bar, 5 mm. C, Average grain filling rate of NILs. D‒G, Comparisons of enzyme activities of adenosine diphosphate glucose (ADPG) (D), granule-bound starch synthase (GBSS) (E), starch branching enzymes (SBE) (F) and soluble starch synthase (SSS) (G) in endosperms of NILs. H‒L, Comparisons of hormone levels of indole-3-acetic acid (IAA) (H), abscisic acid (ABA) (I), cytokinins (CTK) (J), zeatin (ZT) (K) and zeatin riboside (ZR) (L) in endosperms of NILs.Data are Mean ± SD (= 9).
Different lowercase letters above the bars indicate significant differences at< 0.05 determined by the Student- Newman-Keuls test.
Since grain shape, chalkiness degree and filling characteristics all affect taste quality to various degrees (Lestari et al, 2009), comparisons of the nutritional and taste qualities of the NILs were performed. In terms of amylose content, NIL-YF47showed 17.40%, and that of NIL-gw8was significantly reduced to 16.78% (Fig. S2-A), presumably due to the significant reduction in the activities of ADPG, GBSS, SBE and SSS. Similarly, NIL-YF47showed 69.5 mm gel consistency, while NIL-gw8showed a significantly increased gel consistency of 72.2 mm (Fig. S2-B). The protein content and alkali spreading value did not show significant differences among the NILs (Fig. S2-C and -D). Finally, the differences in amylose content and gel consistency significantly affected the taste value (Fig. S2-E), with NIL-gw8achieving a taste value of 72.77, which was significantly higher than that of NIL-YF47(70.66).
Finally, we compared the yield-related traits, including the number of effective panicles per plant, the number of filled grains per panicle and 1000-grain weight, among the NILs in two consecutive growing seasons in 2020 and 2021. In 2020, the number of effective panicles per plant was 14.7 for NIL- YF47, which was significantly lower in NIL-gw8and NIL-gw8(Fig. S3-A). As shown in Fig. S3-B, there was no significant difference in the number of filled grains per panicle among the NILs. In terms of 1000-grain weight, NIL-gw8, NIL-gw8and NIL-gw8were 25.11, 23.01 and 23.11 g,respectively, which were significantly lower than NIL-YF47(26.55 g) (Fig. S3-C). Due to the significant decrease in 1000-grain weight and the number of effective panicles per plant, the yields of gene edited materials were significantly reduced by 7.56%−12.59% compared with NIL-YF47(Fig. S3-D). In 2021, the performance of each yield-related trait was slightly better than that in 2020, whereas the overall trend was similar to that in 2020 (Fig. S3-E to -H).
The application of erect-panicle genefacilitates theformation of high-yielding populations with good ventilation, strong light penetration, and adaptability to dense-planting. However, it also brings the problems of shorter grain shape and slightly higher chalkiness degree due to the uncoordinated strong and weak grain filling (Chen et al, 2012). Therefore, in the context of the widespread use ofin high-yieldingrice, cultivating of rice varieties with longer grain shape and better appearance quality has become a focus issue (Xu et al, 2014; Huang and Qian, 2017). In this study, the different target sites ofediting allcaused inordinately changes in grain shape. Considering the glume cell size and the 1000-grain weight, we speculated that the changes in the glume cells caused by the down-regulation of hormone levels were the main reason for the reduced 1000-grain weight and increased length to width ratio in the NILs, which is similar to the results of previous studies (Li et al, 2018; Mao et al, 2021). Numerous studies showed that chalky endosperm is filled with loosely packed, spherical starch granules, which is mainly caused by the uncoordinated strong and weak grain filling (Wang et al, 2012; Li et al, 2014). All theediting offspring in this study showed a lower grain filling rate compared with NIL-YF47, while the starch-filling enzyme activities and hormone levels were also significantly down-regulated. According to Wang et al (2015), themodule significantly affects the expression of starch biosynthesis genes, and numerous studies have shown that the formation of grain shape, especially grain width, is closely related to grain filling rate (Ishimaru et al, 2013; Wang et al, 2015). Therefore, we speculated that thegene editing may affect the expression of starch biosynthesis genes, thereby causing changes in starch-filling related enzymes and hormones, and ultimately affecting rice grain shape by changing the balance of source-sink flow. In this study, the starch-filling enzyme activities and hormone levels decreased in a large amplitude, and this may be due to the fact that the tested samples were at the peak grain filling stage, which is consistent with the results of our previous study (Mao et al, 2021). The designedmiR156 editing target contains its 20 bp specific sequence, however, mutation occurred at the prior one base of their specific sequence, which may not change the cleavage site of miR156. According to Wang et al (2012), theAmolallele functions as a loss-of-function mutation with slender grains and increased grain yield compared with the plants carrying the ‘Basmati’ allele. Similarly, the grain length, chalkiness and yield of miR156 editing lines were superior than those of the other editing lines in this study. Therefore, we speculated that the editing of the 3rd exon near or in which miR156 is located may cause a stronger loss-of-function type, and the significant down-regulation of gene expression may also partially confirm this speculation.
High yield and superior quality have always been the goals of crop genetic improvement, but it is often difficult to reconcile the two, and grain filling rate may be an important factor to balance yield and appearance quality (Xu and Chen, 2016; Hao et al, 2021; Kan et al, 2022). In this study, originating from a significant decline in grain filling rate, the 1000-grain weight and effective panicle number were significantly decreased, which caused a significant decline in yield of all theediting offspring. Therefore, we speculated that introduction of other grain shape genes on the basis of improvedloci, which are conducive to the increase of grain filling rate, such asor, may be an effective way to balance source-sink flow and create high-yielding and superior-qualityrice. Among theedited materials in this study, NIL-gw8displayed the largest length to width ratio of 2.3, which was significantly different from famousrice known for their long grain shape, such as Daohuaxiang 2 and Jiahe 212, whose aspect ratio reaches about 2.5 (Chen et al, 2018). The mechanism of creating high qualityrice with larger length to width ratio by introducing multiple genes for elongated grain will be the key question of interest.
Taken together, all theediting offspring increased grain length to width ratio by 6%−30%, with a chalkiness degree reduction of 49%−78%. Among them, NIL-gw8showed the highest length to width ratio and the lowest chalkiness degree. In addition, owning to the significant improvement of amylose and gel consistency, the taste quality of NIL-gw8was significantly better than that ofNIL-YF47. Therefore, in the context of rice varieties with abackground, the editing of the miR156 region ofprovided a promising means to select for improved grain qualities inrice, and all the results provided a theoretical basis for the application of manipulating rice functional genes in breeding practices.
ACKNOWLEDGEMENTS
This study was supported by the Central Guidance on Local Science and Technology Development Fund of Liaoning Province, China (Grant No. 2023JH6/100100039), Applied Basic Research Fund of Liaoning Province, China (Grant No. 2022JH2/101300283), National Natural Science Foundation of China (Grant Nos. 32071991 and 32188102), Science Foundationof Liaoning Province, China (Grant No. 2019-ZD-0397), Zhejiang Provincial Natural Science Foundation of China (Grant No. LDQ23C130001), and Key Research and Development Program of Zhejiang Province, China (Grant No. 2021C02056).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
File S1. Methods.
Fig. S1. Basic characteristics of target loci sequences, amino acidvariations,expression variations and plant architecture of near-isogenic lines (NILs).
Fig. S2. Comparisons of nutritional and taste qualities of near-isogenic lines (NILs).
Fig. S3. Comparisons of yield characteristics among near-isogenic lines (NILs).
Chen M J, Liu G F, Yu H, Wang B, Li J Y. 2018. Towards molecular design of rice plant architecture and grain quality., 63: 1276–1289. (in Chinese with English abstract)
Chen W F, Xu Z J, Tang L. 2012. Advances and prospects of rice breeding for super high yield in China., 43(6): 643–649. (in Chinese with English abstract)
Hao J Q, Wang D K, Wu Y B, Huang K, Duan P G, Li N, Xu R, Zeng D L, Dong G J, Zhang B L, Zhang L M, Inzé D, Qian Q, Li Y H. 2021. The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice., 14(8): 1266–1280.
Huang H Y, Qian Q. 2017. Progress in genetic research of rice grain shape and breeding achievements of long-grain shape and good qualityrice., 31(6): 665–672. (in Chinese with English abstract)
Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B I, Onishi A, Miyagawa H, Katoh E. 2013. Loss of function of the IAA-glucose hydrolase geneenhances rice grain weight and increases yield., 45(6): 707–711.
Kan Y, Mu X R, Zhang H, Gao J, Shan J X, Ye W W, Lin H X. 2022.controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis., 8(1): 53–67.
Lestari P, Ham T H, Lee H H, Woo M O, Jiang W Z, Chu S H, Kwon S W, Ma K, Lee J H, Cho Y C, Koh H J. 2009. PCR marker-based evaluation of the eating quality ofrice (L.)., 57(7): 2754–2762.
Li N, Xu R, Duan P G, Li Y H. 2018. Control of grain size in rice., 31(3): 237–251.
Li Y B, Fan C C, Xing Y Z, Yun P, Luo L J, Yan B, Peng B, Xie W B, Wang G W, Li X H, Xiao J H, Xu C G, He Y Q. 2014.encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice., 46(4): 398–404.
Liu Q, Han R X, Wu K, Zhang J Q, Ye Y F, Wang S S, Chen J F, Pan Y J, Li Q, Xu X P, Zhou J W, Tao D Y, Wu Y J, Fu X D. 2018. G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice., 9: 852.
Mao T, Zhu M D, Sheng Z H, Shao G N, Jiao G A, Mawia A M, Ahmad S, Xie L H, Tang S Q, Wei X J, Hu S K, Hu P S. 2021. Effects of grain shape genes editing on appearance quality of erect-panicle geng/rice., 14(1): 74.
Sun S Y, Wang L, Mao H L, Shao L, Li X H, Xiao J H, Ouyang Y D, Zhang Q F. 2018. A G-protein pathway determines grain size in rice., 9: 851.
Tao Y J, Wang J, Xu Y, Wang F Q, Li W, Jiang Y J, Chen Z H, Fan F J, Zhu J P, Li X, Jie Y. 2023. Rational design of grain size to improve rice yield and quality., 30(1): 1−5.
Wang E T, Wang J J, Zhu X D, Hao W, Wang L Y, Li Q, Zhang L X, He W, Lu B R, Lin H X, Ma H, Zhang G Q, He Z H. 2008. Control of rice grain-filling and yield by a gene with a potential signature of domestication., 40(11): 1370–1374.
Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D. 2012. Control of grain size, shape and quality byin rice., 44(8): 950–954.
Wang S K, Li S, Liu Q, Wu K, Zhang J Q, Wang S S, Wang Y, Chen X B, Zhang Y, Gao C X, Wang F, Huang H X, Fu X D. 2015. Theregulatory module determines grain shape and simultaneously improves rice yield and grain quality., 47(8): 949–954.
Xu Q, Xu N, Xu H, Tang L, Liu J, Sun J, Wang J Y. 2014. Breeding value estimation of the application ofandto improvement ofL., 34(4): 1933–1942.
Xu Z J, Chen W F. 2016. Research progress and related problems onsuper rice in northern China., 49(2): 239–250. (in Chinese with English abstract)
Yi C D, Wang D R, Wei J, Li W, Cheng X J, Wang Y, Zhou Y, Liang G H, Gu M H. 2016. Development of functional markers and identification of haplotypes for rice grain shape gene, 42(9): 1297. (in Chinese with English abstract)
December 2022
Copyright © 2023, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2022.12.002
Li Xu (chinalixu1983@163.com);
Hu Shikai (hushikai@caas.cn)
25 October 2022;
19
- Rice Science的其它文章
- Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury
- Transcriptome Analysis of oserf922 Mutants Reveals New Insights into Rice Blast Resistance
- ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice
- LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice
- Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa
- Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth

