Transcriptome Analysis of oserf922 Mutants Reveals New Insights into Rice Blast Resistance
Qin Mengchao, Tao Hui, Shi Xuetao, Zhang Chongyang, He Feng, Wang Min, Liu Zheng, Wang Jisong, Zhang Rongxue, Wang Shutong, Wang Guoliang, Ning Yuese, Wang Ruyi
Letter
Transcriptome Analysis ofMutants Reveals New Insights into Rice Blast Resistance
Qin Mengchao1, 2, #, Tao Hui2, #, Shi Xuetao2, Zhang Chongyang2, He Feng2, Wang Min2, Liu Zheng2, Wang Jisong2, Zhang Rongxue3, Wang Shutong1, Wang Guoliang4, Ning Yuese2, Wang Ruyi2
(College of Plant Protection, Hebei Agricultural University, Baoding 071000, China; State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China; Tianjin Key Laboratory of Crop Genetics and Breeding, Tianjin Academy of Agricultural Sciences, Tianjin 300384, China; Department of Plant Pathology, the Ohio State University, Columbus, OH 43210, USA; These authors contributed equally to this work)
Rice blast disease, caused by fungal,severely threatens food security. The susceptibility () genes have emerged to support pathogenesis, and disruption ofgene usually confers increased resistance to multiple pathogen isolates.Rice ethylene response factor geneis a potentialgene in blast disease. However, hownegatively regulates resistance againstis still unknown.Here, we generatedknockout (Cas9) mutants in Nipponbare background using genome editing technology and found that dysfunction ofenhancedresistance. RNA-Seq analysis identified 2264 up-regulated genes inCas9mutants compared with the wild type at 24 h post-inoculation. qRT-PCR analysis confirmed the inducible expression of several critical immunity pathway-related genes inCas9mutants, suggesting that-mediated blast susceptibility may be related to reactive oxygen species (ROS) production, metabolite accumulation and salicylic acid (SA) signaling.This study provides insight into the roles of thegenein rice immunity.
Rice () is the main source of dietary calories for more than 50% of the world’s population, but rice production is severely affected by rice blast disease caused by the fungus,whichthreatens food security (Wing et al, 2018). Utilizing resistant rice varieties is the most environmentally friendly and sustainable way to control this pathogen.genes, targeted and/or induced by pathogens to support host compatibility and to facilitate infection, are useful targets for breeding resistant varieties (van Schie and Takken, 2014). Indeed, modification or loss of function ofgenes usually confers resistance to related pathogens. For example, disruption of thegene()in rice confers resistance to multipleisolates (Gao et al, 2021). Similarly, knockout of thegene() in wheat confers broad-spectrum resistance to rust fungus (Wang et al, 2022). Disruption ofgenes sometimes results in growth penalties and yield losses. Therefore, breeding resistance materials usinggene should also consider the trade-off between plant growth and immunity. One excellent example is the wheatline, which causes a large deletion in thelocus and shows resistance to powdery mildew without yield loss (Li et al, 2022).
Knocking down the rice ethylene response factor geneinrice cultivar Zhonghua 17 enhances resistance againstisolate P140(Liu et al, 2012). Similarly, a knockout ofproduced by CRISPR/ Cas9-mediated gene editing inrice variety Kuiku 131 increases resistance againstisolate 06-47-6 (Wang et al, 2016). These studies suggested thatacts as a usefulgene in rice blast resistance. However, the mechanism underlying OsERF922-mediated rice immunityremains unclear. To explore the function and regulation ofin blast susceptibility, we used the CRISPR/Cas9 method to specifically target thecoding region (at 134–153 bp downstream from ATG) to generate knockout mutants in therice cultivar Nipponbare (NPB) (Fig. 1-A). We selected two independent homozygous lines 2-1 and 7-1 (with a 1-bp insertion of a T or G base, respectively, leading to early termination of translation) for spray inoculation using the virulentisolate RB22 (Figs. 1-A, -B and S1). As observed in a previous study (Wang et al, 2016), theCas9mutants generated in this study produced fewer disease lesions and accumulated less relative fungal biomass compared with the controlNPB plants (Fig. 1-C to -E).
To explore the regulation of-mediated susceptibility, we performed qRT-PCR assay and found a significant down-regulated expression ofafter inoculated with RB22 compared with 0 h (Fig. S2), implying thatmay be involved in rice andinteraction. We subsequently carried out a transcriptome deep sequencing (RNA-Seq) analysis of NPB andCas9plants before inoculation and at 24 h afterspray inoculation. We identified 104 differentially expressed genes (DEGs) inCas9mutants compared with NPB plants before inoculation, with 73 up-regulated and 31 down-regulated genes (Figs. 1-F and S3), suggesting that knocking outin rice may minimally affect the global transcriptome. Remarkably, 24 h after the onset of inoculation, we detected 5357 DEGs inCas9mutants compared with NPB plants, of which 2264 were up-regulated and 3093 were down-regulated (Figs. 1-F and S3). A Venn diagram illustrated in addition to 3497 co-upregulated DEGs inCas9mutants and NPB plants at 24 h post- inoculation, additional 2440 DEGs were specially induced at 24 h post-inoculation in theCas9mutants, but not in the NPB control (Fig. 1-G), suggesting thatis likely a negative regulator of disease resistance, and these up-regulated genes may attribute to the ability of’s transcription activating function. A Gene Ontology (GO) analysis revealed that the up-regulated DEGs inCas9mutants were mainly associated with the GO categories: protein modification process, metabolic process, adenyl ribonucleotide binding, and transcription factor complex (Fig. S4). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis indicated that the up-regulated DEGs inCas9mutants were over-represented in KEGG pathways related to protein processing in endoplasmic reticulum, plant-pathogen interaction, biosynthesis of cofactor, and MAPK signaling pathway (Fig. S5), which have all been reported to be closely associated with the plant disease resistance responses (Liang and Zhou, 2018; Chen et al, 2020). We concluded thatCas9mutant plants may activate these signaling pathways to execute the immunity response.
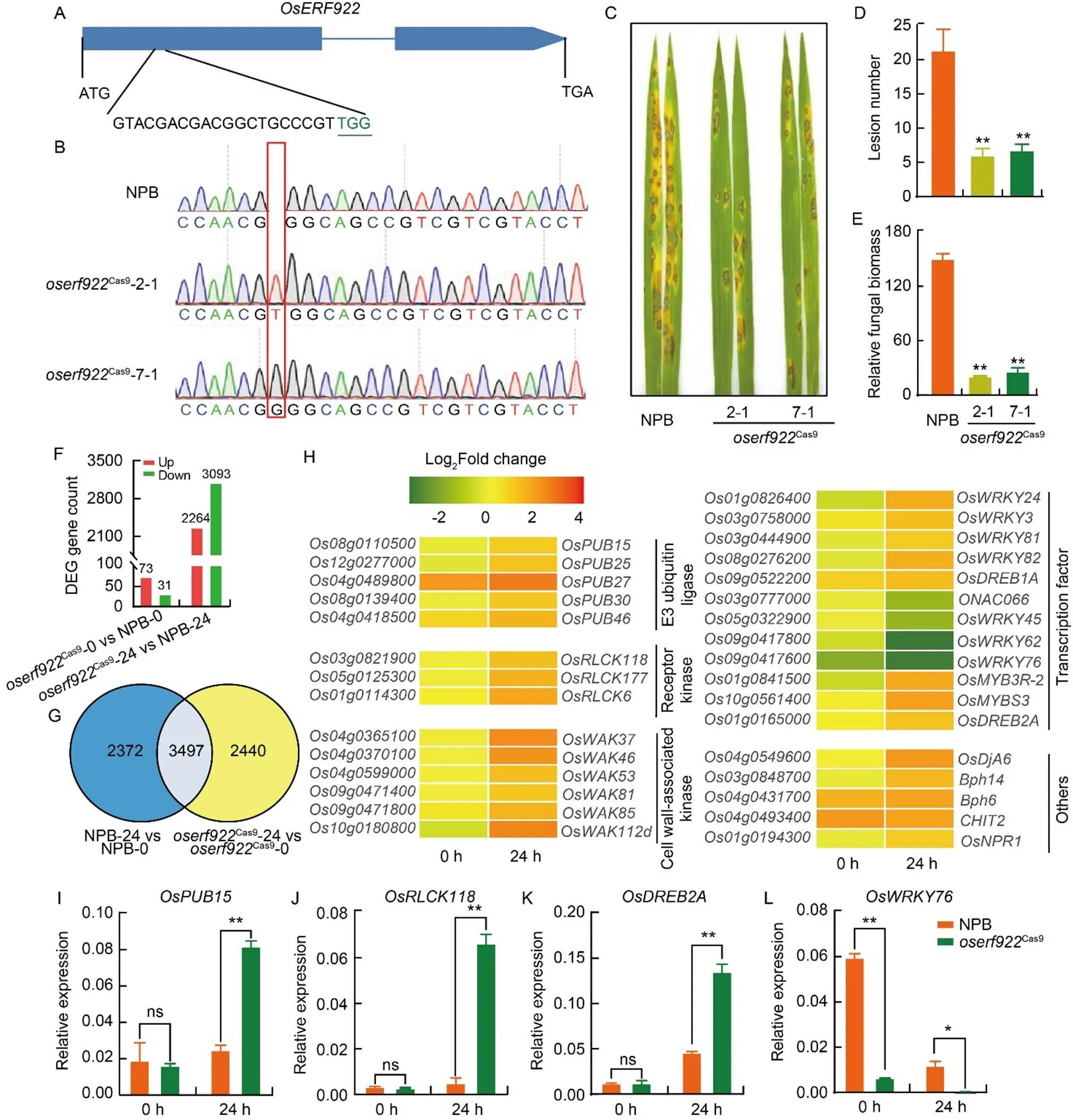
Fig. 1. RNA-Seq analysis of Nipponbare andCas9plants before inoculation and at 24 h post-inoculation with.
A, Diagram ofshowing the position of the sgRNA target site. ATG and TGA represent start and stop codons, respectively. B, Sequence electropherograms ofin wild type Nipponbare (NPB) andCas9mutants. The red rectangle highlights the mutations. C, Representative phenotypes of NPB andCas9mutants following spray inoculation withisolate RB22. D and E, Lesion number (D) and relative fungal biomass (E) of the inoculated leaves in C. F, Analysis of differentially expressed genes (DEGs) of leaves in NPB andCas9mutants before inoculation and at 24 h post-inoculation with RB22. G, Venn diagram showing the extent of overlapping DEGs inCas9mutants and NPB before inoculation and at 24 h post-inoculation with RB22. H, Heatmap representation of the expression levels of selected significantly up-regulated and down-regulated DEGs inCas9mutants and NPB-samples before inoculation and at 24 h post-inoculation with RB22. The genes are classified based on the function of their encoded proteins: E3 ligase, receptor kinase, cell wall-associated kinase, transcription factor, and others. The color scale indicates the expression level of genes from low (green) to high (red). I–L, Relative expression levels of DEGs from the RNA-Seq analysis encoding E3 ligase (I), receptor kinase (J) and transcription factors (K and L), as evaluated by qRT-PCR in leaves of NPB andCas9plants. The internal control gene was.
Data in D, E and I–L are Mean ± SE (= 3). Asterisks indicate statistically significant differences as determined by the Student’s-test (*,< 0.05; **,< 0.01; ns, Not significant).
A growing number of studies have shown that E3 ubiquitin ligases, receptor kinases, cell wall-associated kinases, transcription factors, and others are widely involved in plant immunity (Antolín-Llovera et al, 2012; Duplan and Rivas, 2014; Kohorn, 2016; Wani et al, 2021). Interestingly, we noticed differential expression for such genes inCas9mutant plants compared with NPB plants before inoculation and at 24 h post-inoculation with, including several E3 ubiquitin ligase genes [(,),(), and()], receptor kinase genes [(,),(), and()], cell wall-associated kinase genes [(),(), and()], transcription factor genes [(),(), and()], and others [() and()] (Fig. 1-H).
To verify the accuracy of the RNA-Seq results, we chose several functionally interesting DEGs for qRT-PCR analysis. The E3 ubiquitin ligase OsPUB15 positively regulates blast resistance by inducing the expression of pathogenesis-related genes and accumulation of hydrogen peroxide (Wang et al, 2015). We determined thatexpression was significantly induced at 24 h inoculation in theCas9mutants, but not in NPB plants (Fig. 1-I), indicating that OsERF922 may negatively regulatefollowing blast infection. OsRLCK118 and OsMKK1positively regulate blast resistance, while OsRLCK118 phosphorylates the NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG B (OsRbohB) to promote ROS production (Fan et al, 2018), and OsMKK1promotes ROS accumulation and lipid peroxidation (Dangol et al, 2021). We observed a significant up-regulation ofandat 24 h inoculation in theCas9mutants, but not in NPB plants (Figs. 1-J and S6-A). The induction of,andin theCas9mutants afterinfection suggested that OsERF922-mediated blast susceptibility may be related to ROS accumulation. Over-expression of the transcription factor gene() in soybean () enhances salt tolerance by accumulating soluble sugars and free prolines (Zhang et al, 2013). In this study,was significantly induced by blast infection in both NPB andCas9plants, but to a greater extent in the mutants (Fig. 1-K). OsWRKY76 was reported to be a negative regulator ofresistance (Yokotani et al, 2013). Knockout lines forexhibit an increased accumulation of phytoalexins (Liu et al, 2016). According to the RNA-Seq data,expression was down-regulated byinfection in theCas9mutants compared with the NPB plants. qRT-PCR assay revealed theexpressionwas much lower in theCas9mutants than in the NPB plants before infection (Fig. 1-L), indicating that the transcriptional regulation ofbyOsERF922 may be independent ofinfection. Moreover,expression further decreased after pathogen infection (Fig. 1-L). A diterpenoid phytoalexins synthase gene(Cho et al, 2004)was significantly induced in theCas9mutants afterinfection (Fig. S6-B). The differential expression of,andin theCas9mutants suggested that OsERF922-mediated blast susceptibility may be related to the accumulation of metabolites. SA is an essential hormone in plant immunity, and over-expression of SA signaling transcriptional regulator encoding geneenhances resistance to the rice blast fungus (Feng et al, 2011).is induced in response to SA analog benzothiadiazole andinfection (Reyna and Yang, 2006). In this study,andwere significantly induced after blast infection in theCas9plants compared with the NPB plants (Fig. S6-C and -D). The up-regulation expression level of these genes indicated that-mediated blast susceptibility may be involved in SA signaling and biosynthesis.
OsERF922 is a transcriptional activator belonging to the APETELA2/ERF-type transcription factor family that regulates the transcription of downstream target genes by binding to the GCC box sequence in their promoters (Liu et al, 2012). Our transcriptome data revealed that several critical resistance- related genes were up-regulated in themutants (Figs. 1-I, -J, and S6-C, -D). The promoter sequences of these DEGs will be analyzed in the future to investigate potential direct target genes of OsERF922. We believed that these findings may offer valuable candidate genes for genetic manipulation of disease resistance in crop breeding improvement.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (Grant Nos. U20A2021 and 32161143009) and Tianjin Natural Science Foundation, China (Grant No. 19JCZDJC34200).
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science; http://www.ricescience.org.
File S1. Methods.
Fig. S1. Predicted protein sequence of OsERF922 in Nipponbare andCas9mutants.
Fig. S2. Expression ofin Nipponbare before and afterinfection evaluated by qRT-PCR assay.
Fig. S3. Differential gene expression analysis at 0 and 24 h post-inoculation.
Fig. S4. Gene Ontology (GO) term enrichment analysis of differentially expressed genes at 24 h post-inoculation.
Fig. S5. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of differentially expressed genes at 24 h post-inoculation.
Fig. S6. qRT-PCR assay confirmed expression of differently expressed genes in Nipponbare (NPB) andCas9plants before and afterinfection.
Antolín-Llovera M, Ried M K, Binder A, Parniske M. 2012. Receptor kinase signaling pathways in plant-microbe interactions., 50: 451–473.
Chen Q, Yu F F, Xie Q. 2020. Insights into endoplasmic reticulum- associated degradation in plants., 226(2): 345–350.
Cho E M, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K J, Oikawa H, Toshima H, Shibuya N, Nojiri H, Omori T, Nishiyama M, Yamane H. 2004. Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor., 37(1): 1–8.
Dangol S, Nguyen N K, Singh R, Chen Y F, Wang J, Lee H G, Hwang B K, Jwa N S. 2021. Mitogen-activated protein kinase OsMEK2 and OsMPK1 signaling is required for ferroptotic cell death in rice-interactions., 12: 710794.
Duplan V, Rivas S. 2014. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity., 5: 42.
Fan J B, Bai P F, Ning Y S, Wang J Y, Shi X T, Xiong Y H, Zhang K, He F, Zhang C Y, Wang R Y, Meng X Z, Zhou J G, Wang M, Shirsekar G, Park C H, Bellizzi M, Liu W D, Jeon J S, Xia Y, Shan L B, Wang G L. 2018. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice., 23(4): 498–510.
Feng J X, Cao L, Li J, Duan C J, Luo X M, Le N, Wei H H, Liang S J, Chu C C, Pan Q H, Tang J L. 2011. Involvement ofin rice basal resistance to blast fungus., 131(2): 221–235.
Gao M J, He Y, Yin X, Zhong X B, Yan B X, Wu Y, Chen J, Li X Y, Zhai K R, Huang Y F, Gong X Y, Chang H Z, Xie S H, Liu J Y, Yue J X, Xu J L, Zhang G Q, Deng Y W, Wang E T, Tharreau D, Wang G L, Yang W B, He Z H. 2021. Ca2+sensor- mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector., 184(21): 5391–5404.
Kohorn B D. 2016. Cell wall-associated kinases and pectin perception., 67(2): 489–494.
Li S N, Lin D X, Zhang Y W, Deng M, Chen Y X, Lv B, Li B S, Lei Y, Wang Y P, Zhao L, Liang Y T, Liu J X, Chen K L, Liu Z Y, Xiao J, Qiu J L, Gao C X. 2022. Genome-edited powdery mildew resistance in wheat without growth penalties., 602: 455–460.
Liang X X, Zhou J M. 2018. Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling., 69: 267–299.
Liu D F, Chen X J, Liu J Q, Ye J C, Guo Z J. 2012. The rice ERF transcription factor OsERF922 negatively regulates resistance toand salt tolerance., 63(10): 3899–3911.
Liu J Q, Chen X J, Liang X X, Zhou X G, Yang F, Liu J, He S Y, Guo Z J. 2016. Alternative splicing of riceandtranscription factor genes in pathogen defense., 171(2): 1427–1442.
Reyna N S, Yang Y N. 2006. Molecular analysis of the rice MAP kinase gene family in relation toinfection., 19(5): 530–540.
van Schie C C N, Takken F L W. 2014. Susceptibility genes 101: How to be a good host., 52: 551–581.
Wang F J, Wang C L, Liu P Q, Lei C L, Hao W, Gao Y, Liu Y G, Zhao K J. 2016. Enhanced rice blast resistance by CRISPR/ Cas9-targeted mutagenesis of the ERF transcription factor gene., 11(4): e0154027.
Wang J, Qu B Y, Dou S J, Li L Y, Yin D D, Pang Z Q, Zhou Z Z, Tian M M, Liu G Z, Xie Q, Tang D Z, Chen X W, Zhu L H. 2015. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity., 15: 49.
Wang N, Tang C L, Fan X, He M Y, Gan P F, Zhang S, Hu Z Y, Wang X D, Yan T, Shu W X, Yu L G, Zhao J R, He J N, Li L L, Wang J F, Huang X L, Huang L L, Zhou J M, Kang Z S, Wang X J. 2022. Inactivation of a wheat protein kinase gene confers broad- spectrum resistance to rust fungi., 185(16): 2961–2974.
Wani S H, Anand S, Singh B, Bohra A, Joshi R. 2021. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects., 40(7): 1071–1085.
Wing R A, Purugganan M D, Zhang Q F. 2018. The rice genome revolution: From an ancient grain to Green Super Rice., 19(8): 505–517.
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H, Kaku H, Minami E, Nishizawa Y. 2013. WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance., 64(16): 5085–5097.
Zhang X X, Tang Y J, Ma Q B, Yang C Y, Mu Y H, Suo H C, Luo L H, Nian H. 2013., a rice transcription factor, significantly affects salt tolerance in transgenic soybean., 8(12): e83011.
Copyright © 2023, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2023.05.002
Wang Ruyi (wangruyi@caas.cn);
Tao Hui (htao2018@126.com)
4 April 2023;
24 May 2023
- Rice Science的其它文章
- Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury
- Effect of GW8 Gene Editing on Appearance Quality of Erect-Panicle Type (dep1) Japonica Rice
- ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice
- LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice
- Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa
- Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth

