rs10795668单核苷酸多态性与中国北方汉族结直肠癌易感性的关系
冯凯,路浩军,贾海英,郑燕华,张军,景青萍,吴继华
rs10795668单核苷酸多态性与中国北方汉族结直肠癌易感性的关系
冯凯,路浩军,贾海英,郑燕华,张军,景青萍,吴继华
目的探讨rs10795668单核苷酸多态性与北方汉族人群结直肠癌遗传易感性的关系。方法采用聚合酶链反应-限制性片断长度多态性方法,检测结直肠癌组182例(结肠癌113例,直肠癌69例)和对照组192例的rs10795668单核苷酸多态性基因型。采用非条件logistic回归分析进行基因-疾病关联分析与基因-性别交互作用分析。结果rs10795668多态性位点在结直肠癌患者中的基因型分布与在正常健康对照人群中的基因型分布差异无统计学意义,但基因型与性别在结肠癌中存在交互作用,AA+AG基因型增加女性结肠癌的发生危险(OR=3.05,95%CI 1.47~6.35;P=0.00078)。结论rs10795668单核苷酸多态性的AA+AG基因型与中国北方汉族女性结肠癌遗传易感性有关,与直肠癌无关。
单核苷酸多态性;结直肠肿瘤;遗传易感性
结直肠癌(colorectal cancer,CRC)是全球发病率最高的肿瘤之一[1],每年约有120万人罹患结直肠癌,并导致60万人死亡[2]。病因学上,多种遗传和环境因子可导致结直肠癌的发生[3-6]。
基因组计划完成以来,在候选基因策略研究中虽然发现了一些风险遗传位点,但可重复、功能确定的位点依然有限。近来,全基因组关联研究迅猛发展,已报道了多个常见的大肠癌易感遗传位点,包括5q31.1,8q23.3,8q24.21,10p14,10q22.3,10q25.2,11q12.2,11q23.1,12p13.31,14q22.3,15q13.3,16q22.1,18q21.1,17p13.3,19q13.2、20p12.3等[7-15]。在上述位点中,位于10p14的单核苷酸多态性(SNP)rs10795668是最早发现的风险因子之一[12]。大量的重复研究显示,rs10795668在不同族群乃至同一族群中的结果并不一致[16-18]。针对多数东亚人群包括中国人群的研究显示rs10795668与结直肠癌易感性有关[19-20],这与欧洲人群相似,而与非洲人群不同。本研究采用182例结直肠癌患者与192例对照进行rs10795668的关联重复研究,并对解剖位置分层,进行了性别与基因的交互作用分析。
1 材料与方法
1.1 研究对象 所有病例均来自2011-2014年解放军306医院普通外科与肿瘤科收治的手术或化疗的北方汉族结直肠癌患者,病理组织类型均为原发腺癌,182例结直肠癌病例中包括113例结肠癌(男63例,女50例,年龄64.6±14.1岁)与69例直肠癌(男40例,女29例,年龄67.0±11.9岁),选取192例与病例组年龄、性别比例接近的北方汉族健康体检人员做对照。为排除遗传分层因素,所有入选者均经详细问卷调查,以查明其疾病史并排除有肿瘤病史者。样本采用EDTA抗凝全血。本研究经解放军第306医院伦理委员会认可。
1.2 基因型分析 采用酚-氯仿法提取结直肠癌患者与正常对照者外周血白细胞基因组DNA,-70℃保存备用。限制性片段长度多态性方法设计采用在线工具SNP-RFLPing2(http://bio.kuas.edu.tw/snp-rflping2/ rflpUI.jsp.)。参考序列为(chr10:8700719-8701719),版本号为GRCh37/hg19。引物序列如下:上游引物5'-TGAGGCTTATTGAAAACAATGG-3',下游引物5'-GCAGAGGTTGCATGCAGTG-3'。采用TaKaRa公司RCR试剂盒进行PCR反应。反应体系为25μl,PCR反应条件为:94℃5min;94℃30s,58℃30s,72℃30s,共16个循环;94℃30s,56℃退火40s,72℃40s,20个循环,最后72℃10min,4℃保存。以上反应在GeneAmp PCR system 9700型(美国PE应用生物系统公司)基因扩增仪上进行。PCR产物大小为293bp,限制性内切酶为AluⅠ,完全酶切产物为180bp和114bp两个片段。PCR反应后取产物10μl,采用限制性内切酶进行酶切,37℃水浴,过夜。酶切产物用2%琼脂糖凝胶电泳,并以ChampGel全自动凝胶成像分析仪观察记录电泳结果。对三种基因型各取5例进行测序证实(北京三博远志生物工程公司)。
1.3 rs10795668 SNP的基因组注释和连锁不平衡(linkage disequilibrium,LD)分析 基因组注释采用UCSC基因组浏览器(http://genome.ucsc.edu/cgibin/hgGateway),参考版本号为GRCh37/hg19。LD分析采用SNAP网络工具(http://www.broadinstitute. org/mpg/snap/ldplot.php),参数设置选择Regional LD Plots,SNP data set设为1000 genomes pilots,Population panel设为CHBJPT,r2设为0.6,Distance limit设为100kb。
1.4 统计学处理 用χ2检验分析对照组人群基因型分布是否符合Hardy-Weinberg(H-W)平衡,以判断其对整体人群的代表性。基因疾病关联分析采用非条件logistic回归模型,计算共显性、显性、隐性、超显性遗传模式下的比数比(odds ratios,OR)及95%可信区间(confidential intervals,CI)。对疾病类型与性别进行分层,并采用logistic回归模型进行基因-性别交互作用分析,所有统计皆采用SNPStats在线关联研究工具进行分析[21]。以上检验显著性水准均采用α=0.05。
2 结 果
2.1 一般特征 结直肠癌组与对照组之间性别、年龄构成相似(P>0.05,表1)。所有标本均成功进行了基因型分型(图1),对部分结果进行了测序验证,所有重复分型结果均与原结果相符。χ2检验显示,健康对照组rs10795668基因型分布符合H-W平衡(P=0.75)。
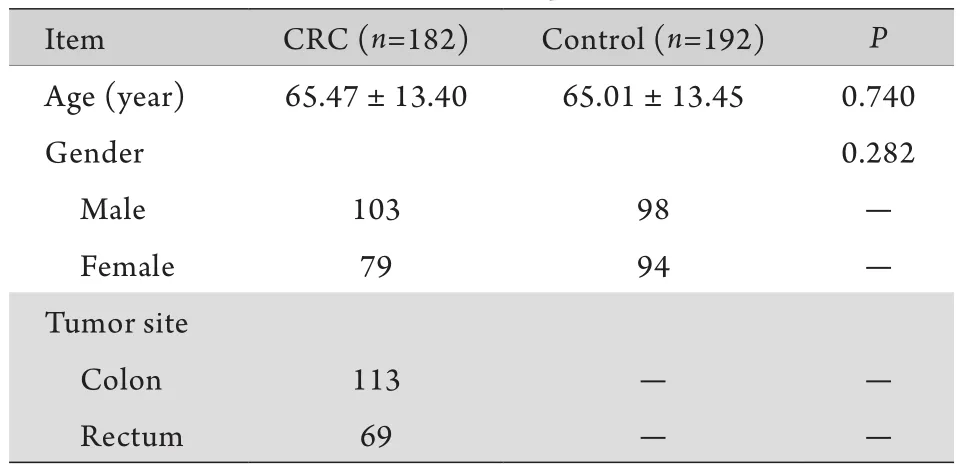
表1 结直肠癌组与对照组性别、年龄分布及肿瘤部位Tab.1 Distributions of sex and age among CRC patients and controls, and the tumor site in CRC patients
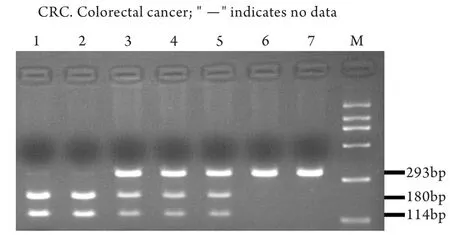
图1 rs107595668基因多态性分析Fig.1 Genotype analysis of rs10795668 polymorphismM. DL2000 DNA ladder; Lane1-2. A/A homozygous (180bp and 114bp); Lane 3-5. G/A heterozygous (293bp, 180bp and 114bp); Lane 6-7. G/G homozygous (293bp)
2.2 rs10795668 SNP与结直肠癌及结、直肠癌亚组的关联研究 rs10795668 SNP的G/G、G/A和A/A基因型频率在结直肠癌组与对照组之间,各种遗传模式下差异均无统计学意义(表2)。将结肠癌和直肠癌患者分成两个亚组分别与对照组进行关联分析在四种遗传模式下差异亦无统计学意义(数据未列出)。以上分析均校正了年龄和性别因素。

表2 rs10795668单核苷酸多态性与结直肠癌的关联分析(n=374)Tab.2 Association analysis of rs10795668 with CRC (n=374)
2.3 rs10795668 SNP在结直肠癌及结、直肠癌亚组中的基因-性别交互作用研究 结直肠癌rs10795668 SNP与性别的交互作用采用logistic回归模型进行分析,结果显示,在显性遗传模式下,结直肠癌rs10795668 SNP与性别存在交互作用,携带A等位基因(G/A+A/A)的女性患结直肠癌比不携带A等位基因的女性的高(OR=1.81,95% CI 1.00~3.29,P=0.0089,表3),在其他三种遗传模式下未观察到交互作用(数据未提供)。为了观察这种交互作用是否与结直肠癌亚型有关,分别进行了结、直肠癌亚组中的基因-性别交互分析,结果显示,显性遗传模式下,rs10795668 SNP与性别存在交互作用主要体现在结肠癌亚组(OR=3.05,95% CI 1.47~6.35,P=0.00078,表4),直肠癌亚组没有显著交互作用(表5),即其他条件相同的情况下携带A等位基因的女性更易发生结肠癌。以上分析均校正了年龄因素。
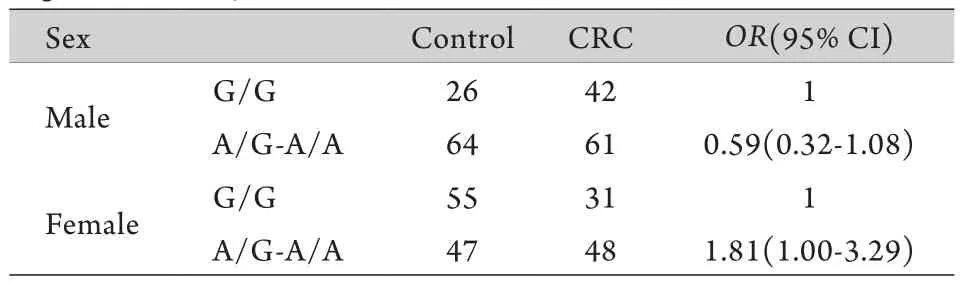
表3 Logistic回归分析结直肠癌基因和性别的交互作用(n=374)Tab.3 Interaction of gene-sex involved in CRC (logistic regression analysis,n=374)

表4 Logistic回归分析结肠癌基因和性别的交互作用(n=305)Tab.4 Interaction of gene-sex involved in colon cancer (logistic regression analysis,n=305)
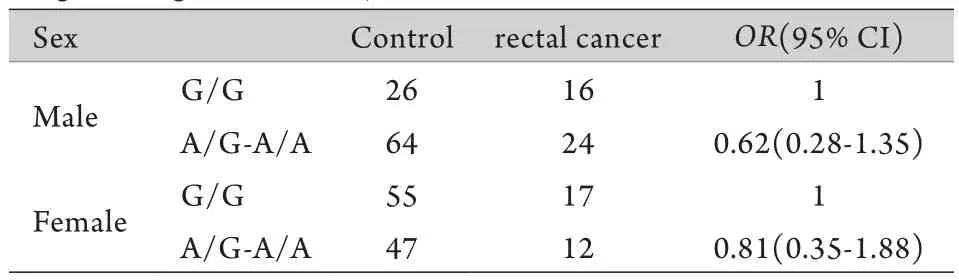
表5 Logistic回归分析直肠癌基因和性别的交互作用(n=261)Tab.5 Interaction of gene-sex involved in rectal cancer (logistic regression analysis,n=261)
2.4 rs10795668 SNP基因组注释与LD分析
rs10795668位于10号染色体8701219位(图2),LD范围54.3kb(r2≥0.6,图3),距离rs10795668最近的基因是编码tRNA的RNA5SP299(2.4kb)。最近的编码蛋白的基因是GATA3,此为重要的转录因子,距rs10795668位点585.2kb。

图2 UCSC基因组浏览器所示rs10795668参考序列Fig.2 The UCSC genome browser view of chr10: 8,000,000-9,000,000 (build 37) containing rs10795668The red rectangle represents susceptibility SNP rs10795668. The UCSC Gene track shows multiple transcripts span around rs10795668. The nearest gene is a tRNA gene,RNA5SP299. The blue underline show 2 lincRNA genes located in 200kb upstream and downstream, respectively. The nearest coding protein gene is GATA3, underlined by purple line. The light blue rectangle shows the reported eQTL gene, ATP5C1 (Loo LW, 2012)
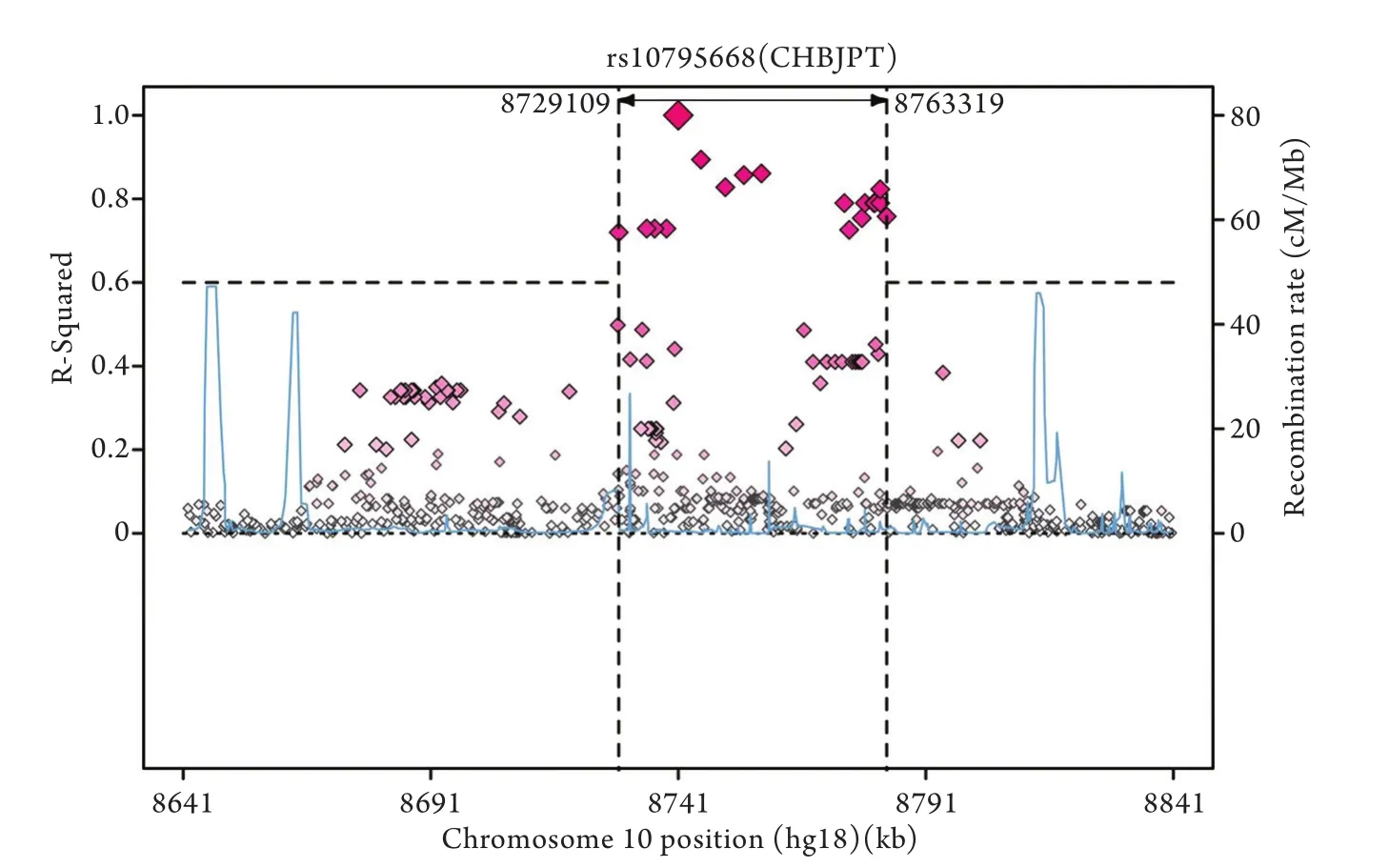
图3 rs107595668的LD范围示意图(r2≥0.6)Fig. 3 Regional LD plot for SNPs rs107595668 at 10p14, associated with colorectal cancer (r2≥0.6)
3 讨 论
本研究结果显示,rs10795668SNP多态位点在结直肠患者中的基因型分布与正常对照样本差异无统计学意义,但基因型与性别在结肠癌中存在交互作用,A等位基因型增加了中国北方汉族女性结肠癌的发生危险。
rs10795668SNP与结直肠癌关联的报道最早见于2008年在欧洲人群中的一项GWAS研究[12],随后在不同人群中进行了多个重复研究工作,其中有很多矛盾的报道[15-24]。rs10795668人群分布差异很大,千人基因组计划数据(http:// browser.1000genomes.org/index.html)显示,rs10795668 A等位基因在欧洲人群约为32%,东亚洲人群为37%,西亚人群为22%,非洲人群为2%。通常报道,在欧洲和亚洲G为易感等位基因,而在非洲A为易感等位基因。因此,种族因素与结直肠癌易感性密切相关。虽然最近有Meta分析显示,在中国人群中,rs10795668 SNP与结直肠癌相关[17],且G等位基因增加结肠癌的发病风险,但本研究没有重复出这样的结果,且在进一步对样本进行分层分析和基因型与性别的交互作用研究中,发现A等位基因极为显著地增加女性结肠癌的发病风险,这在其他研究中尚未见报道。结肠癌与直肠癌一般统称结直肠癌,但这两者的发病机制可能存在差异[25],并且性别可能是影响其易感性的重要因素之一[26]。
rs10795668并不位于某一基因附近,所以其功能研究是个难点。生物信息学分析显示,rs10795668位于10号染色体8701219位,LD范围54.3kb(r2≥0.6)。最近的编码蛋白的基因是GATA3,它是个重要的转录因子。目前只有1 篇rs10795668功能研究的报道[27],该报道显示rs10795668等位基因与ATP5C1的表达水平相关。但是,由于ATP5C1与rs10795668的相对距离较远,间隔数个转录子,包括GATA3和多个lincRNA转录子,这些都是重要的调控因子,特别是lincRNA近来是研究调控靶基因表达的热点[28-29],因此ATP5C1表达水平是否真与rs10795668等位基因关联还需要进一步确认。另外,rs10795668作为遗传标记的 LD范围为54.3kb(r2≥0.6),在其关联范围内,就只有编码tRNA的RNA5SP299基因,但目前关于tRNA与基因多态性的研究还鲜有报道。
本研究对结直肠癌患者样本分层,并与性别和rs10795668不同等位基因进行交互作用研究,提示A等位基因增加中国北方汉族女性结肠癌的易感性,相关机制有待进一步探索。
[1]Xin L, Bai Y, Li ZS. Advances in colorectal cancer risk factors[J]. Chin J Pract Intern Med, 2014, 34(12): 1214-1218. [辛磊, 柏愚,李兆申. 结直肠癌危险因素研究进展[J]. 中国实用内科杂志, 2014, 34(12): 1214-1218.]
[2]Jemal A, Bray F, Center MM,et al. Global cancer statistics[J]. CA Cancer J Clin, 2011, 61(2): 69-90.
[3]de la Chapelle A. Genetic predisposition to colorectal cancer[J]. Nat Rev Cancer, 2004, 4(10): 769-780.
[4]Lichtenstein P, Holm NV, Verkasalo PK,et al. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland[J]. N Engl J Med, 2000, 343(2): 78-85.
[5]Wang H, Fu ZX, Lu WD,et al. Effect and mechanism of Prdx2 on proliferation and apoptosis of colorectal cancer cell line SW480[J]. Med J Chin PLA, 2014, 39(4): 293-296. [王昊, 傅仲学, 卢伟东, 等. Prdx2在结直肠癌细胞增殖、凋亡中的作用及其机制研[J]. 解放军医学杂志, 2014, 39(4): 293-296.]
[6]Wang RH, Xie JG, Fu Q,et al. Expression of Ang-2, Tie-2 mRNA and K-ras gene mutation in human colorectal cancer tissue[J]. J Zhengzhou Univ(Med Sci), 2013, 48(5): 629-632. [王瑞华, 谢建国, 付强, 等. 结直肠癌组织中Ang-2、Tie-2 mRNA的表达及K-ras基因突变的检测[J]. 郑州大学学报(医学版), 2013, 48(5): 629-632.]
[7]Zanke BW, Greenwood CM, Rangrej J,et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24[J]. Nat Genet, 2007, 39(8): 989-994.
[8]Tomlinson I, Webb E, Carvajal-Carmona L,et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21[J]. Nat Genet, 2007, 39(8): 984-988.
[9]Broderick P, Carvajal-Carmona L, Pittman AM,et al. A genomewide association study shows that common alleles of SMAD7 influence colorectal cancer risk[J]. Nat Genet, 2007, 39(11): 1315-1317.
[10] Jaeger E, Webb E, Howarth K,et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk[J]. Nat Genet, 2008, 40(1): 26-28.
[11] Tenesa A, Farrington SM, Prendergast JG,et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21[J]. Nat Genet, 2008, 40(5): 631-637.
[12] Tomlinson IP, Webb E, Carvajal-Carmona L,et al. A genomewide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3[J]. Nat Genet, 2008, 40(5): 623-630.
[13] Houlston RS, Cheadle J, Dobbins SE,et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33[J]. Nat Genet, 2010, 42(11): 973-977.
[14] Dunlop MG, Dobbins SE, Farrington SM,et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk[J]. Nat Genet, 2012, 44(7): 770-776.
[15] Jia WH, Zhang B, Matsuo K,et al. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer[J]. Nat Genet, 2013, 45(2): 191-196.
[16] Kupfer SS, Anderson JR, Hooker S,et al. Genetic heterogeneity in colorectal cancer associations between African and European americans[J]. Gastroenterology, 2010, 139(5): 1677-1685, 1685 e1671-1678.
[17] Qin Q, Liu L, Zhong R,et al. The genetic variant on chromosome 10p14 is associated with risk of colorectal cancer: results from a case-control study and a meta-analysis[J]. PLoS One, 2013, 8(5): e64310.
[18] Wang H, Haiman CA, Burnett T,et al. Fine-mapping of genomewide association study-identified risk loci for colorectal cancer in African Americans[J]. Hum Mol Genet, 2013, 22(24): 5048-5055.
[19] Hong SN, Park C, Kim JI,et al. Colorectal cancer-susceptibility single-nucleotide polymorphisms in Korean population[J]. J Gastroenterol Hepatol, 2015, 30(5): 849-857.
[20] Zhang B, Jia WH, Matsuda K,et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk[J]. Nat Genet, 2014, 46(6): 533-542.
[21] Sole X, Guino E, Valls J,et al. SNPStats: a web tool for the analysis of association studies[J]. Bioinformatics, 2006, 22(15): 1928-1929.
[22] Middeldorp A, Jagmohan-Changur S, van Eijk R,et al. Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort[J]. Cancer Epidemiol Biomarkers Prev, 2009, 18(11): 3062-3067.
[23] Li FX, Yang XX, Hu NY,et al. Single-nucleotide polymorphism associations for colorectal cancer in southern chinese population[J]. Chin J Cancer Res, 2012, 24(1): 29-35.
[24] Talseth-Palmer BA, Brenne IS, Ashton KA,et al. Colorectal cancer susceptibility loci on chromosome 8q23.3 and 11q23.1 as modifiers for disease expression in Lynch syndrome[J]. J Med Genet, 2011, 48(4): 279-284.
[25] Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer[J]. Nature, 2012, 487(7407): 330-337.
[26] Feng K, Lu HJ, Jia HY,et al. Association between the single nucleotide polymorphism of RASSF1A Ala133Ser and colorectal cancer in north Chinese Han population[J]. Med J Chin PLA, 2015, 40(3): 221-225. [冯凯, 路浩军, 贾海英, 等. RASSF1A A la133Ser单核苷酸多态性与中国北方汉族结直肠癌的关联研究[J]. 解放军医学杂志, 2015, 40(3): 221-225.]
[27] Loo LW, Cheng I, Tiirikainen M,et al. cis-Expression QTL analysis of established colorectal cancer risk variants in colon tumors and adjacent normal tissue[J]. PLoS One, 2012, 7(2): e30477.
[28] Roux BT and Lindsay MA. LincRNA signatures in human lymphocytes [J]. Nat Immunol, 2015, 16(3): 220-222.
[29] Ma W, Ay F, Lee C,et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes[J]. Nat Methods, 2015, 12(1): 71-78.
Relationship of the polymorphism of rs10795668 single nucleotide and the susceptibility of colorectal cancer in north China Han population
FENG Kai1, LU Hao-jun1, JIA Hai-ying2, ZHENG Yan-hua1, ZHANG Jun1, JING Qing-ping3, WU Ji-hua3*1Center for Special Medicine and Experimental Research,2Center for Medical Examination,3Department of Pathology, 306 Hospital of PLA, Beijing 100101, China
*< class="emphasis_italic">Corresponding author, E-mail: jh_02821@126.com
, E-mail: jh_02821@126.com
This work was supported by the Medical Research Grant of 306 Hospital of PLA (13QN01;13ZD11)
ObjectiveTo evaluate the relationship between the rs10795668 single nucleotide polymorphism (SNP) and the colorectal cancer (CRC) in north China Han population.MethodsThe genotype of rs10795668 SNP was detected by PCRRFLP analysis in 113 colon cancer patients, 69 rectal cancer patients, and 192 age and sex matched controls. Unconditional logistic regression were used to comparatively analyze the genotype between patients and the controls adjusted by age and sex.ResultsNo significant difference of rs10795668 SNP was detected between colorectal cancer patients and normal controls, but it was found that an existence of interaction of gene and sex in colorectal cancer, and women with AA+AG genotype may have lower risk of colon cancer (OR3.05, 95% CI 1.47-6.35;P=0.00078).ConclusionThe AA+AG genotype of rs10795668 SNP is associated with the genetic susceptibility of colon cancer, but not of rectal cancer, in north China Han women.
colorectal cancer; genetic susceptibility; single nucleotide polymorphism
R735.34
A
0577-7402(2015)10-0821-05
10.11855/j.issn.0577-7402.2015.10.10
2015-06-01;
2015-08-29)
(责任编辑:沈宁)
解放军306医院院级课题(13QN01;13ZD11)
冯凯,医学博士,副主任医师,副教授。主要从事重大疾病遗传易感性方面的研究
100101 北京 解放军第306医院特种医学实验研究中心(冯凯、路浩军、郑燕华、张军),体检中心(贾海英),病理科(景青萍、吴继华)
吴继华,E-mail:jh_02821@126.com

