DNA End Configurations Regulate RecJ Exonuclease Activity*
MA Lan‑Zhi,XIE Yan,ZHANG Peng,CHEN Hou‑Zao,LIU De‑Pei
(State Key Laboratory of Medical Molecular Biology,Department of Biochemistry and Molecular Biology,Institute of Basic Medical Sciences,Chinese Academy of Medical Sciences&Peking Union Medical College,Beijing 100005,China)
Abstract Objective DNA end resection is a common mechanism for the formation of 3'‑ssDNA tails in homologous recombination (HR) and is mainly mediated by 5'‑3' exonuclease. However, whether DNA end configurations directly regulate 5'‑3'exonuclease activity remains unclear.In this study,we explored the regulation and mechanisms of DNA end configurations on RecJ,the only 5'‑3' exonuclease of RecF recombination pathway in Escherichia coli. Methods To investigate the regulation of DNA 3'‑end configurations on RecJ exonuclease, single‑stranded DNAs (ssDNAs) containing different lengths of 3'‑ssDNA overhangs (9 nt and 18 nt) and 3'‑end modifications (phosphorylation and phosphorothioation) were used for exonuclease assays in the presence of Mg2+.To elucidate the mechanisms, RecJ was incubated with substrates containing different 3'‑end configurations in the absence of Mg2+and analyzed by electrophoretic mobility shift assays(EMSA).Furthermore,the coordination of DNA 3'end configurations and two other RecJ regulatory factors,DNA 5'‑end phosphorylation and single‑stranded DNA binding protein(SSB),were determined by exonuclease assays and EMSA on substrates with different 3'‑end configurations respectively. Results DNA 3'‑end configurations inhibited the RecJ exonuclease activity, including DNA 3'‑overhang length and 3'‑end modifications (phosphorylation and phosphorothioation). 3'‑End phosphorylation and phosphorothioation of DNA reshaped the RecJ‑DNA binding patterns to inhibit RecJ exonuclease activity. DNA 5'‑end phosphorylation overcame RecJ inhibition of 3'‑end modifications and remodeled the RecJ‑DNA binding patterns. In addition, SSB partially overcame the 3'‑end modifications mediated inhibition by enhancing RecJ‑DNA binding.Conclusion The RecJ exonuclease activity was regulated and orchestrated by the DNA configurations of 3'and 5'ends.
Key words homologous recombination,RecJ exonuclease,DNA end configuration,DNA end resection regulation
DNA is constantly attacked by both endogenous and exogenous deleterious agents. DNA double‑strand breaks(DSBs)are among the most dangerous types of DNA damage, thus, DSBs must be repaired to ensure genomic stability and integrity. Homologous recombination (HR) and non‑homologous end‑joining(NHEJ) are two distinct pathways for repairing DSBs[1]. HR‑mediated repair has relatively high fidelity, whereas NHEJ is error‑prone. In mammalian cells, DSBs are predominantly repaired by NHEJ[2].However, wild‑type Escherichia coli relies heavily on two HR pathways, RecBCD and RecF, for adequate DSBs repair[3‑5].
HR‑mediated repair initiates with DNA end processing, which generates long 3' single‑stranded DNA (ssDNA) tails for the following steps[6]. Due to the diverse configurations of broken DNA ends,various nucleases and other accessory factors are required to ensure the uniform generation of long 3'‑ssDNA tails, such as 5'‑3' exonucleases, which trim DNA in the 5'‑3' direction[7]. As the only 5'‑3'exonuclease in the RecF pathway[5,8‑9], RecJ resects diverse DNA ends to create 3'‑ssDNA tails. Thus, the mechanisms by which RecJ adapts to distinct DNA ends are vital for fine‑tuning the DNA end processing.
RecJ prefers ssDNA substrates and requires metal ions for DNA trimming[8‑10]. In fact, RecJ cannot degrade substrates effectively in the absence of Mg2+or Mn2+[8‑10]. RecJ also resects double‑stranded DNA (dsDNA) with 5′‑ssDNA overhangs[11‑12].Similarly, drRecJ, a RecJ ortholog of Deinococcus radiodurans, shows a preference to digest hairpin DNA with 5'‑ssDNA overhangs. This substrate specificity shows that RecJ activity can be directly regulated by the 5' ends of DNA. In fact, it has been shown that the length and modifications of 5'‑ssDNA tails can regulate the RecJ exonuclease activity[11,13‑15].However, the regulatory effects of 3'‑end configurations on the RecJ exonuclease activity have remained unclear.
Consequently, we investigated the regulation of length and modifications of 3'‑ssDNA overhangs on RecJ exonuclease activity and then we elucidated the mechanisms. We found that DNA 3'‑end configurations, including the 3'‑ssDNA‑overhang length and 3'‑end modifications, inhibited RecJ exonuclease activity. On the contrary, the 5'‑end phosphorylation and single‑stranded DNA binding protein (SSB) partially overcame the 3'‑end modifications caused inhibition. Based on the above,we revealed that DNA end configurations regulate RecJ exonuclease activity, and it is plausible that such end‑configuration‑mediated exonuclease activity coordination is a common mechanism for all organisms.
1 Materials and methods
1.1 Enzymes and proteins
RecJ (M0264L, 3×107U/L) used in this study was obtained from New England Biolabs (NEB,USA), which is a recombinant of the native enzyme fused with maltose binding protein(MBP)and has the same enzymatic properties as wild‑type RecJ.Extreme thermostable SSB (M2401S, 0.5 g/L) was obtained from NEB.
1.2 DNA detection reagents
The ssDNA 20 ladder was purchased from Biolular (B1001). SYBR Green II RNA gel stain was purchased from Thermo Fisher Scientific (S7568).The chemiluminescent nucleic acid detection module kit was purchased from Beyotime(D3308).
1.3 DNA substrates
Four hairpin‑structure ssDNAs were designed with an 8‑bp stem, 3' tails of 9 or 18 nt, and 5' ends starting with thymine or guanine, which were nominated as T‑8‑9, T‑8‑18, G‑8‑9, and G‑8‑18,respectively (Figure 1a). The sequence of T‑8‑18 was derived from the previous research[16], and other DNA sequences with or without modifications were listed in Table S1 in Supplementary. In particular,phosphorothioate (PS) modified DNA sequences contain 5 phosphorothioate bonds. All DNA oligonucleotides were synthesized by Sangon Biotech(Shanghai,China).
1.4 Annealing of ssDNA substrates
DNA substrates were annealed by heating at 95℃for 5 min and slowly cooling to 4℃before reacting with RecJ and SSB.
1.5 Exonuclease activity
To determine the metal dependence of RecJ exonuclease, 2 μmol/L substrates (T‑8‑9 and T‑8‑18)were incubated with 90 U RecJ in the presence or absence of 10 mmol/L Mg2+.
For a typical RecJ exonuclease assay, RecJ exonuclease reactions (50 μl) were performed with 2 μmol/L DNA substrates containing different 3'‑end configurations and 90 U RecJ in a solution of 50 mmol/L NaCl, 10 mmol/L Tris‑HCl (pH 8.0),1 mmol/L 1, 4‑dithiothreitol (DTT), and 10 mmol/L MgCl2. The reactions were incubated for 2 h at 37℃followed by 65℃for 20 min to inactivate the RecJ,and then loaded onto denaturing urea‑PAGE gels composed of 8% acrylamide, 7 mol/L urea, and 0.5×Tris‑borate‑ethylenediaminetetraacetic acid (EDTA)(TBE) buffer. Gels were pre‑run in 0.5×TBE buffer;then, the samples were loaded and electrophoresed in 0.5×TBE buffer at 4℃and 120 V for 60 min. The gels were stained with SYBR Green II RNA gel stain in 0.5×TBE buffer at 25℃for 40 min. Fluorescence signals were visualized using a Typhoon FLA 9500 scanner(GE Healthcare).
RecJ exonuclease time gradient (1 or 2 h) tests were performed with 2 μmol/L substrates containing different 3'‑end modifications in the presence of 10 mmol/L Mg2+.
For RecJ exonuclease activity assays with SSB,2 μmol/L DNA substrates were prebound with a SSB gradient (0-20 mg/L) for 30 min. Subsequently, 90 U RecJ was added to the binding reactions and incubated at 37℃for 2 h.
1.6 Electrophoretic mobility shift assays
To analyze the interaction between RecJ and different ssDNA substrates, a reaction mixture (20 μl)containing 10 mmol/L Tris‑HCl (pH 7.5), 1 mmol/L DTT, 50 mmol/L KCl, 50% glycerol, 25 mmol/L EDTA, 5 nmol/L biotin‑labeled ssDNA, and RecJ gradient(0-570 U)was incubated at 30℃for 30 min,which was similar to the previous study[8], and followed by incubation at 4℃for 2 h. The samples were then loaded onto native gels composed of 8%acrylamide and 0.5×TBE buffer. Gels were pre‑run in 0.5×TBE buffer. Samples were electrophoresed in 0.5×TBE buffer at 4℃and 80 V for 90 min and then transferred to a nylon membrane in 0.5×TBE buffer at 4℃and 250 mA for 40 min. Biotin signals were detected using a chemiluminescent nucleic acid detection module kit (Beyotime, D3308), according to the manufacturer’s instructions.
For the electrophoretic mobility shift assays(EMSA) with SSB, DNA substrates (10 nmol/L) were incubated with SSB(10 mg/L)for 90 min at 30℃in a binding reaction buffer (20 μl) containing 10 mmol/L Tris‑HCl (pH 7.5), 1 mmol/L DTT, 50 mmol/L KCl,50% glycerol, and 25 mmol/L EDTA. For supershift reactions, 570 U RecJ was added to the binding reaction mixture containing 10 nmol/L DNA and 500 nmol/L SSB.
1.7 RecJ concentration determination
The molar concentration of RecJ was quantified by bovine serum albumin (BSA) standard curve.Standard curve was fitted by BSA gradients (0-250 mg/L). Proteins were boiled, and then supplied to SDS‑PAGE and Coomassie bright blue staining. Gray statistics were analyzed by ImageJ(Version 1.52a).
1.8 Statistical analysis
The significance of the differences between pairs of groups was determined using an unpaired t‑test(Welch’s t‑test). Differences among the three groups were estimated using Brown‑Forsythe and Welch ANOVA tests with Dunnett’s T3 multiple comparisons. Data are presented as mean±SEM. Data processing and analyses were conducted by ImageJ(Version 1.52a). Statistical significance was set at P <0.05, and all statistical tests were two‑sided.Significance was defined as *P<0.05, **P<0.01, and***P<0.001.
2 Results
2.1 3'-End configurations of ssDNAs inhibit the RecJ exonuclease activity
We first confirmed the metal dependence of RecJ exonuclease using substrates with 3'‑ssDNA overhangs (T‑8‑9 and T‑8‑18) in the presence or absence of 10 mmol/L Mg2+. The results showed that Mg2+activated the RecJ to degrade T‑8‑9 and T‑8‑18(Figure S1a in Supplementary), which is consistent with previous studies[8‑9,13].
Then,we investigated the effect of the 3'‑ssDNA‑overhang length on the RecJ exonuclease activity in the presence of Mg2+and found that RecJ degradation abilities depended on the length of DNA 3' tails. The exonuclease activity of RecJ was significantly weaker in the substrates with 18‑nt 3'tails(T‑8‑18)than in the substrates with 9‑nt 3'‑ssDNA overhangs (T‑8‑9)(Figure 1b, e), indicating that the RecJ exonuclease activity was inhibited by the length of the 3'‑ssDNA overhangs.
We also evaluated the ability of RecJ to degrade substrates with different 3'‑end modifications. The 3'‑phosphorylated terminal is a common form of 3'‑blocking damage caused by oxidative stress damage[17], and phosphorothioate (PS) modification is a well‑known modification that blocks nuclease cleavage[18]. So, we performed exonuclease assays on substrates which 3' ends were modified with either a phosphate group (3'P) or 5 phosphorothioate groups(3' PS). Interestingly, both the 3' P and 3' PS of T‑8‑9 and T‑8‑18 substrates inhibited RecJ from degrading them. The 3' PS substrates showed stronger inhibition than the 3'P substrates(Figure 1c,d,f,g).
To validate the inhibitory effect of DNA 3'‑end modifications on RecJ, we also detected the RecJ exonuclease degradation of substrates containing different 3'‑end modifications at different time points(1 or 2 h). The results showed that the 2 h RecJ incubation led to more substrates degradation, and the inhibition of DNA 3'‑end modifications on RecJ was more pronounced at 2 h than that at 1 h(Figure S1b-e in Supplementary).
Since RecJ is a 5'‑3'exonuclease,G‑8‑9 and G‑8‑18 were used to explore whether the DNA 5'‑end base pairs affect the inhibition of RecJ exonuclease by DNA 3'‑end configurations. 3'‑ssDNA‑overhang length and 3'‑end modifications of G‑8‑9 and G‑8‑18 showed similar inhibition to their A‑T counterparts with comparable degradation rates (Figure S2 in Supplementary). Therefore, the inhibition of the RecJ exonuclease activity by the 3'‑end configurations was not affected by the 5'‑end base pairs. These results revealed that the 3'‑end configurations,including the 3'‑ssDNA‑overhang length and 3'‑end modifications,inhibit the RecJ exonuclease activity.
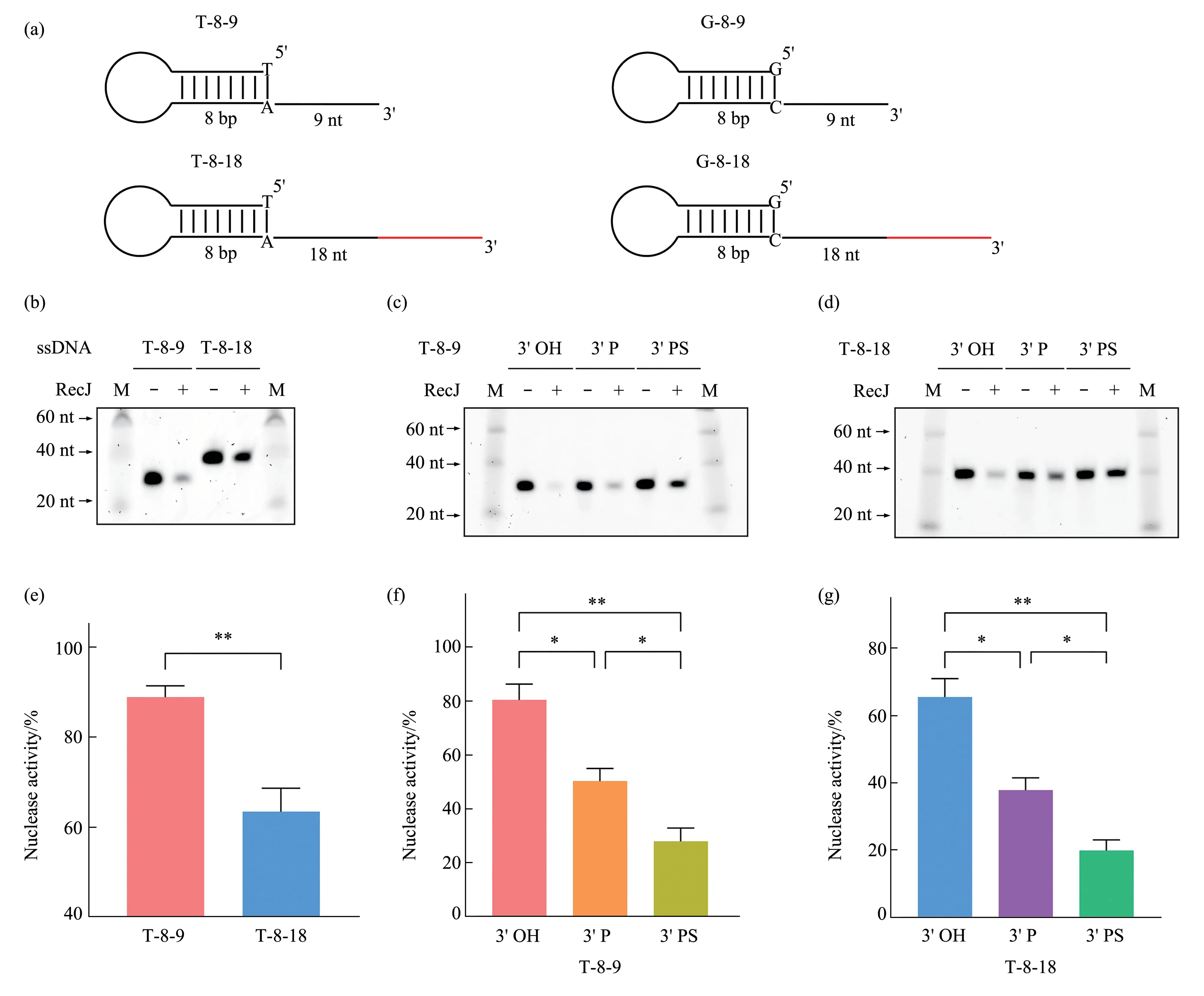
Fig.1 3'-End configurations of ssDNAs inhibit the RecJ exonuclease activity(a) Schematic diagram of DNA substrates, the length as well as type of ends for each DNA is indicated. (b) RecJ exonuclease activity on substrates with different 3'‑ssDNA‑overhang length. T‑8‑9 and T‑8‑18 (2 μmol/L) were incubated with RecJ (90 U) in the presence of 10 mmol/L Mg2+ and detected using urea‑PAGE gel staining.(c,d)RecJ exonuclease activity on ssDNAs with different 3'‑end modifications:T‑8‑9(c)and T‑8‑18(d)with 3'‑end phosphorylation(3'P)or phosphorothioation(3'PS)(2 μmol/L)were incubated with RecJ(90 U)in the presence of 10 mmol/L Mg2+and then detected using urea‑PAGE gel staining. (e-g) Quantification of the RecJ exonuclease activity on ssDNA substrates with different 3'‑end configurations (n=4 independent replicate experiments). Data are represented as mean±SEM. P values were calculated using Welch’s t‑test (e) and Brown‑Forsythe and Welch ANOVA tests(f-g).M,marker.*P <0.05,**P <0.01.
2.2 3'-End configurations modulate the RecJDNA binding
To clarify the mechanisms by which the 3'‑end configurations inhibit the RecJ exonuclease activity,we performed EMSA to test the binding of RecJ to DNA substrates with different 3'‑end configurations in the absence of Mg2+. To optimize the binding of RecJ and ssDNA, we first quantified RecJ concentration by BSA standard curve (Figure S3a, b in Supplementary). The concentration of MBP‑RecJ was (55.97±3.03) mg/L (n=4), hence, the molar concentration of RecJ was about 550 nmol/L.We then incubated ssDNA substrates (T‑8‑18 and T‑8‑18‑P)with RecJ gradient (0-500 nmol/L) in the absence of Mg2+to determine the RecJ‑DNA binding capacity.The results showed that the RecJ‑DNA binding ability was relatively weak, but the RecJ‑DNA binding was enhanced with the increase of RecJ concentration(Figure S3c, d in Supplementary), which is consistent with previous studies[8,13‑14]. So we used 500 nmol/L RecJ (570 U) for the following RecJ‑DNA binding tests.
Surprisingly, the 3'‑end modifications did not hinder the binding ability of RecJ but rather changed its binding patterns. Figure 2 showed that the binding of RecJ to DNA substrates was relatively weak.However,the presence of more than one binding band indicates that RecJ and the substrate (T‑8‑18) have different binding states.Both the 3'P and 3'PS altered the RecJ‑DNA binding patterns. Specifically, the 3' P and 3'PS promoted the formation of RecJ‑DNA binding complex with low shift ability and slightly hindered the formation of binding complex with high shift ability.
We also investigated the effect of the 3'‑ssDNA‑overhang length on the RecJ‑DNA binding. EMSA were performed with RecJ and substrates (T‑8‑9 and T‑8‑18)in the absence of Mg2+.Although there was no distinct difference in the binding bands between the two groups, T‑8‑18 had a lower binding ratio (bound/free)and a less substrate loss than T‑8‑9(Figure S4 in Supplementary), indicating that the length of the 3'‑ssDNA overhangs may hinder the RecJ binding affinity to substrates.

Fig.2 3'-End modifications of ssDNAs remodel the RecJ-DNA binding patternsBiotin‑labeled ssDNA substrates (5 nmol/L) with different 3'‑end modifications including 3' P and 3' PS: T‑8‑18 and T‑8‑18‑P (a), T‑8‑18 and T‑8‑18‑PS(b)were incubated with 570 U RecJ in the absence of Mg2+.The formed complexes were separated using native PAGE gels,and the biotin signals were detected.“P”represents phosphate group;“PS”represents phosphorothioate;“”represents biotinylated T.
2.3 Phosphorylation of 5' ends enhances the RecJ exonuclease activity and overcomes the 3'-end modifications inhibition
The 5'‑phosphate binding pocket of RecJ which is located above the active site determines the 5'‑3'DNA binding polarity of RecJ[8]. Recently, Cheng et al.[14]have shown that the 5'‑end phosphorylation of 20 nt poly (dA) moderately enhances the binding affinity and catalytic efficiency to Deinococcus radiodurans RecJ (drRecJ). We found that the 5'‑end phosphorylation of hairpin ssDNAs enhanced the RecJ exonuclease activity on the 3'‑ssDNA overhangs(Figure 3a-d). To further elucidate whether the 5'‑end phosphorylation can overcome the inhibitory effect of the 3'‑end configurations on RecJ activity, we compared the RecJ exonuclease activity in 3'‑end‑modified substrates with or without 5'‑end phosphorylation. As expected, the 5'‑end phosphorylation largely enhanced the RecJ digestion of substrates with 3'‑end modifications, including 3' P and 3' PS (Figure 3e-l). These findings showed that the phosphorylation of the 3' and 5' ends oppositely regulate the RecJ exonuclease activity, which may provide a coordination mechanism in vivo to fine‑tune DNA end processing in DSB repair.
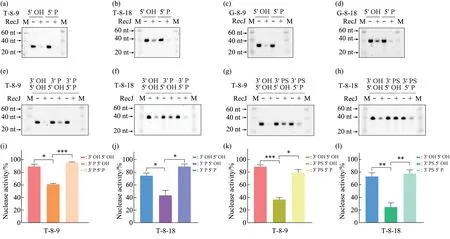
Fig.3 Phosphorylation of 5'ends enhances the RecJ exonuclease activity and overcomes the 3'-end modifications inhibition(a-d) Different ssDNA substrates:T‑8‑9 (a),T‑8‑18 (b), G‑8‑9 (c), and G‑8‑18 (d) (2 μmol/L) with 5' phosphorylated (5' P) or unphosphorylated (5'OH)ends,as shown at the top of the panel,were incubated with RecJ(90 U)in the presence of Mg2+and then detected using urea‑PAGE gel staining.(e-h)RecJ exonuclease activity on different 3'‑end‑modified substrates with 5'P or 5'OH:(e,f)T‑8‑9 and T‑8‑18 with or without 3'P and 5'P;(g,h)T‑8‑9 and T‑8‑18 with or without 3'PS and 5'P.(i-l)Quantification of the RecJ exonuclease activity on different ssDNA substrates(n≥3 independent replicate experiments). Data are represented as mean±SEM. P values were calculated by Brown‑Forsythe and Welch ANOVA tests. M, marker.*P <0.05,**P <0.01,***P <0.001.
2.4 Phosphorylation of 5' ends modulates the RecJ-DNA binding patterns to orchestrate RecJ activity
We used EMSA to elucidate the mechanisms of the 5'‑end phosphorylation to facilitate the RecJ exonuclease activity. RecJ was incubated with ssDNA substrates with 18‑nt 3' tails, with or without 5'‑end phosphorylation (P‑T‑8‑18 and T‑8‑18)in the absence of Mg2+. Unexpectedly, the 5'‑end phosphorylation did not significantly enhance the binding of RecJ to substrates but rather remodeled their binding patterns (Figure 4a). Compared to T‑8‑18, the RecJ and P‑T‑8‑18 binding complex with high shift ability was reduced, whereas the binding complex with low shift ability was increased.Furthermore, the 3'‑end‑modified substrates (T‑8‑18‑P and T‑8‑18‑PS) showed similar remodeling of the RecJ‑DNA binding patterns (Figure 4b, c). Thus, the 5'‑end phosphorylation may enhance the RecJ exonuclease activity by modulating its binding states in a configuration‑dependent manner.
2.5 SSB enhances RecJ degradation and binding
SSB has been shown to promote the RecJ exonuclease activity[8,11‑13]. We found that SSB enhanced the RecJ digestion of the 3'‑end‑modified ssDNAs (Figure S5a-d in Supplementary), and also overcame the RecJ inhibition induced by 3'‑end modifications (Figure S5e-l in Supplementary).EMSA showed that adding SSB resulted in a supershift band and a reduction in free substrates(Figure S6a, b in Supplementary). Therefore, SSB may enhance the RecJ exonuclease activity by promoting its binding to ssDNAs.
3 Discussion
DNA end processing is crucial for providing long 3'‑ssDNA tails for HR‑mediated DSB repair.Nucleases and other accessory factors generate 3'‑ssDNA tails from diverse DNA ends.As the only 5'‑3'exonuclease in the RecF pathway[8‑9], RecJ is versatile and participates in mismatch repair[19‑20], ssDNA gap repair, and base excision repair[14,21]. Previous studies have shown that RecJ digests hairpin DNA and dsDNA with 5'‑ssDNA overhangs but does not trim DNA with blunt ends or 3'‑ssDNA overhangs[8,10‑11,13].However, we observed that RecJ degraded DNA substrates with 3'‑ssDNA overhangs, and the RecJ exonuclease activity was strongly affected by the DNA end configurations. Specifically, both the length and end modifications (3' P and 3' PS) of the 3'overhangs inhibited RecJ digestion (Figure 1, Figure S1, S2 in Supplementary). Analogously, previous studies have shown that the length and modifications of 5'‑ssDNA tails also regulate the RecJ exonuclease activity[10‑11,13‑15]. Moreover, we found that the DNA 5'‑end phosphorylation enhanced RecJ degradation(Figure 3a-d), which is consistent with previous founding that phosphorylation of DNA 5' end enhanced the drRecJ catalytic efficiency[14].Furthermore, we found that the DNA 5'‑end phosphorylation partially overcame the inhibition caused by the 3'‑end modifications(Figure 3e-l).
As opposed to previous reports showing that RecJ binds to ssDNA substrates homogeneously[8,13],we determined that RecJ formed more than one bound complex with ssDNAs containing 3' overhangs(Figure 2, 4).As RecJ source and quality are possible concerns, we quantified RecJ concentration by BSA standard curve (Figure S3a, b in Supplementary).The concentration of MBP‑RecJ was (55.97±3.03)mg/L (n=4), hence, the molar concentration of RecJ was about 550 nmol/L. Nevertheless, neither the 3′‑end modifications nor the 5'‑end phosphorylation significantly affected RecJ binding to ssDNAs but rather reshaped their RecJ‑DNA binding patterns(Figure 2, 4). In addition, based on the reduction of binding ratio and the substrate loss (Figure S4 in Supplementary), it is reasonable that the length of the 3'‑ssDNA overhangs inhibits the RecJ exonuclease activity by affecting its DNA‑binding affinity. Similar RecJ‑DNA binding changes were obtained when using a different gel recipe from Han et al.[13], which reveals the robustness of the RecJ‑DNA binding patterns. Thus, we showed for the first time that RecJ binds to ssDNAs heterogeneously and that DNA end configurations modulate the RecJ‑DNA binding patterns.
RecJ has been widely used as a powerful tool in vitro due to its 5'‑3' exonuclease activity. RecJ is applied to remove the failed chains generated in oligonucleotide synthesis or the excess linkers during library preparation[15‑16,22]. Therefore, our work suggests that the specificity of RecJ can be improved by manipulating the DNA end configurations, further broadening the application potential of RecJ.
Additionally, we found that SSB could enhance RecJ binding to 3'‑end‑modified ssDNAs, and the supershift band indicated the formation of the RecJ‑ssDNA‑SSB complex (Figure S6a, b in Supplementary).SSB is pivotal in the replication and DNA repair processes[23‑24]. Therefore, SSB diminution of the RecJ inhibition triggered by DNA 3'‑end modifications provides another regulatory level to the DNA end processing in vivo.As the RecF pathway is highly conserved[25], we presume that such end‑configuration‑mediated regulation of nuclease activity may be a common mechanism, but how these resection factors and intertwined proteins are orchestrated in the DNA context is unclear and requires further research.
4 Conclusion
RecJ exonuclease degrades DNA substrates with 3'‑ssDNA overhangs. Both DNA 3'‑end and 5'‑end configurations directly regulate RecJ exonuclease activity. DNA 3'‑end and 5'‑end phosphorylation coordinate to regulate RecJ exonuclease activity.
Acknowledgements We would like to thank Editage(www.editage.cn)for English language editing.
Supplementary PIBB_20220131_Doc_S1.pdf is available online (http://www.pibb.ac.cnorhttp://www.cnki.net).

