Whole Genome Sequencing and Comparisons of Different Chinese Rabies Virus Lineages Including the First Complete Genome of an Arctic-like Strain in China*
LI Hao, GUO Zhen Yang, ZHANG Jian, TAO Xiao Yan, ZHU Wu Yang, TANG Qing, and LIU Hong TuKey Laboratory for Medical Virology, Ministry of Health, National Institute for Viral Disease Control and Prevention Chinese Center for Disease Control and Prevention, Beijing 102206, China
Original Article
Whole Genome Sequencing and Comparisons of Different Chinese Rabies Virus Lineages Including the First Complete Genome of an Arctic-like Strain in China*
LI Hao, GUO Zhen Yang, ZHANG Jian, TAO Xiao Yan, ZHU Wu Yang, TANG Qing#, and LIU Hong Tu#
Key Laboratory for Medical Virology, Ministry of Health, National Institute for Viral Disease Control and Prevention Chinese Center for Disease Control and Prevention, Beijing 102206, China
Abstract
Objective To learn the rabies genome molecular characteristics and compare the difference of China rabies lineages.
Methods The complete genomes of 12 strains from different China rabies lineages were amplified and sequenced, and all the China street strain genomes (total 43), Arctic and Arctic-like genomes were aligned using ClustalX2, the genome homologies were analyzed using MegAlign software, and the phylogenetic trees were constructed by MEGA 5.
Results First Arctic-like rabies genome in China (CQH1202D) was reported, and we supplemented the rabies genome data of China, ensuring at least one genome was available in each China lineage. The genome size of China V (11908nt) is obviously shorter than other lineages' (11923-11925nt) for the difference of N-P non-coding regions. Among different lineages, the genome homologies are almost under 90%. CQH1202D (China IV lineage) has close relationship with strains from South Korea and they share about 95% genome similarities.
Conclusion The molecular characteristics of 6 different China rabies lineages were compared and analyzed from genome level, which benefits for continued comprehensive rabies surveillance, rabies prevention and control in China.
Rabies virus; Genome; Lineage
www.besjournal.com (full text) CN: 11-2816/Q Copyright ©2016 by China CDC
INTRODUCTION
Rabies in China represents an ongoing threat to public health and has been a notifiable communicable disease since 1950, with three major rabies epidemics periods documented since this time[1-2]. The disease is predominantly found in the southern provinces of China, with very few cases occurring in northern China[3]. As a consequence of prevention and control efforts, the human incidence has been decreasing since 2007, as evidenced by significant drop in annual human cases in traditional epidemic provinces[4-5]. However, several other provinces,mainly in northern and western China, have experienced continued emergence or reemergence of rabies after several years without any human rabies cases[6-7].
Except a bat Lyssavirus strain, Irkut Virus (first found in Russia), was isolated in Northeast China in 2012[8], Rabies virus (RABV), mainly circled in dogs, is the only viral agent responsible for the severe rabies situation in China[2-3]. Furthermore, RABV in China were divided into 6 lineages according to the comprehensive phylogenetic analysis based on almost all the street strains sequenced in China[2,9],and the two lineages (China I and China II) included almost all the strains, especially for China I, most of the strains collected in the provinces with increasing rabies cases in recent years belong to China I[2,6]. However, in Qinghai and Tibet, where no rabies cases were reported for many years, China IV lineage (corresponding to Arctic-like-2 cluster in world rabies)strains were confirmed, and which showed that at least two rabies origins influenced current China rabies epidemic[7]. So, domestic animals (mainly dogs)play a pivotal role in spreading the disease, and meanwhile wildlife animals (ferret badger, wolf and fox) play a potentially important role in spreading rabies in recent years[7].
In this study, for the first time the whole genome sequence of an Arctic-like rabies virus recovered in China was reported. Furthermore,different genomes belonging to district lineages were compared, to learn the rabies genome molecular characteristics in China.
MATERIALS AND METHODS
Ethics Statement
The program for collection of dog and cow brain specimens was approved by the Ethical Committee of the National Institute of Viral Disease Control and Prevention, China CDC. Dog and cow brain specimens were taken after suspected rabies death.
Epidemiological Data
Data on human rabies cases for each province from the whole of Mainland China were collected from the annual reports of the China CDC. The reporting methods and how cases were determined to be associated with rabies were the same as described previously[5,10].
Specimen Detection
The dog and cow brain tissues were analyzed using the direct immunofluorescence assay (DFA) as described previously[11]. All the background data on specimens were provided by provincial or local CDCs.
RT-PCR and Sequencing
Total RNA extraction and cDNA synthesis procedures were the same as described previously[6]. The complete genomes of the 12 strains were amplified and sequenced with primers designed according to the alignment results of almost all the rabies virus genomes public in GenBank[12]. The 3' end and 5' end of the genomes were confirmed using a 3' and 5' Full RACE Kit (TaKaRa Biotechnology Co., Ltd.,Dalian, China) according to the manufacturer's instructions, and the sequencing results were checked and assembled as described previously[12].
Homology and Phylogenetic Analysis
Complete genomes alignment were performed using ClustalX2[13]. MegAlign software version 5 was used to analyze genome homologies. Phylogenetic reconstruction was performed using MEGA ver. 5.2 with the Neighbor-Joining (NJ) method and branch support estimated using bootstrap sampling of 1000 replicates[14]. Reference sequences that were representative of the major lineages both globally and within China were selected from GenBank based on the earlier studies.
RESULTS
China Rabies Strains Newly Sequenced and Cited
In the previous studies, we classified almost all the Chinese rabies strains into six lineages based on their N or G nucleotide sequences[2,9]. To better know the molecular characteristics of different lineages, several rabies strains belong to the different lineages were selected and sequenced the complete genomes. Among them, 9 strains (CJS0621D, CJS0636D, CJS0848D, CJX0903D,CJX0906D, CSD0708D, CSX0904D, CNM1101C,CNM1104D) belonged to China I, the current most important lineage; CHN0802D was one of China II lineage; CNM1103C was from China III lineage;CQH1202D was the first China IV (corresponding to Arctic-like clade) genome[7]. The newly or recently sequenced 12 rabies genomes in this study are with accession numbers JQ970480-JQ970487,KC193267, KC252633, KC252634, and KM272192. The geography distribution of theses strains are showed in Figure 1.
Except for the newly sequenced 12 genomes, all the other rabies genomes of Chinese street strains published in GenBank were cited and analyzed. Although, no new China V lineage strains werecollected in our lab, the genomes of two old strains (J & CQ92) have been reported[15]. In addition, our lab already reported previously one China VI lineage strain CYN1109D genome[12].
Now the whole genomes of different Chinese rabies lineages were available, so we can compare and analyze the molecular characteristics of different lineages.
Phylogenetic Analysis of China Rabies Street Strains Genomes
Totally, 43 genomes of Chinese rabies street strains were used to built the phylogenetic tree (Figure 2): 27 genomes belonged to China I lineage; 8 strains were China II lineage; 4 genomes were classified China III; China V included two strains; China IV and VI each only have one genome.
Genome Homology Comparison of Different China Rabies Lineages
Genome homology analysis shows that all the Chinese rabies strains shares 83%-99.8% genome similarities. The genome similarities in lineage China I, China II, China III, and China V are respectively 93.1%-99.8%, 92.7%-99.2%, 90%-99.1%, and 99.2% (only one strain in China IV and China VI). Among the different lineages, the genome homologies are almost under 90% (Table 1).
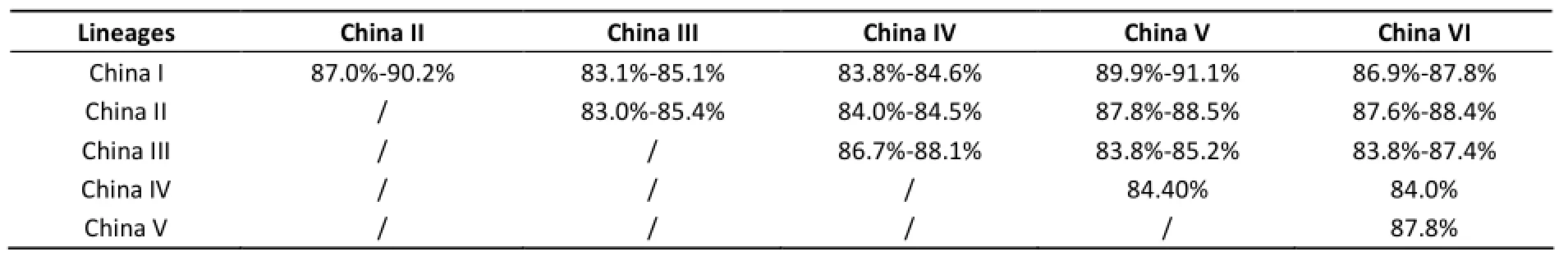
Table 1. The Genome Homology Similarities Between Each Two Different Lineages
Genome Structure of Different Chinese Rabies Lineages
The size of non-coding regions, coding regions andwhole genome of 43 Chinese strains (Figure 2) were calculated respectively, and were showed in Table 1 as different lineages. The 5'UTR of DRV and MRV strains are not complete, so the size of 5'UTR and genome of the two strains are not included in Table 2.
The size of 5 coding regions (N, P, M, G, L)are completely identical in each China lineages;among these lineages, only L coding region is differentin length, the size of China III and IV (6384nt) are shorter than other 4 lineages(6387nt). Relative to the coding regions, the non-coding regions are variable except the conserved 3'UTR. The N-P non-coding regions of China V lineage (73nt) are much shorter than other lineages' (90 or 91nt),which results the difference of genome size between China V (11908nt) and other lineages (11923-11925nt).
Comparison of Arctic/Arctic-like Rabies Genomes
Almost all the Arctic and Arctic-like rabies genomes were cited to construct the phylogenetic tree with CQH1202D strain (Figure 3). China strain CQH1202D shows close relationship with wild animal strains from South Korea; however, the strains isolated from South Asia (Pakistan, India, Bangladesh)are different obviously. Arctic and Arctic-like group is divided with Cosmopolitan, Asian, Indian subcontinent, and American groups.
Genome homology analysis is coincident with above phylogenetic results of Arctic and Arctic-like clade. CQH1202D has 94.8%-95% genome similarities with three South Korea strains,91.4%-91.7% with 6 South Asia strains, only 90.9% with the Russia strain.

Table 2. Non-coding Regions, Coding Regions and Genomes Size (in nucleotides) of Different China Rabies Lineages
DISCUSSION
Facing the more and more serious rabies epidemic in China, China government started the national rabies surveillance program in 2005[5]. More than 10,000 animal (mainly include dogs) and patient samples were collected and detected, and near 400 specimens were identified rabies positive by our lab (national rabies surveillance lab)[2,5]. To learn the characteristics of China rabies lineage distribution,almost all the Chinese rabies street strains were analyzed and divided into 6 China lineages (China I-VI) according to their complete nucleotide sequences of N or G gene[2,9-10]. Different lineage has different population size and geographic distribution[2,7,9]. China I is the most prevalent and dominant lineage in China, and almost all the provinces, except ones in Northeast China and northwest China, were occupied and even keep expanding further. China II has the second large size and distribution, remain constrained to the southwestern provinces. China III-VI only include few strains from one or two provinces. However, the tiny lineage China IV (corresponding to Arctic-like clade in the world) began to occur in Qinghai and Tibet in 2012, was responsible for the wild animal rabies epidemic in these regions[7].
So, it's very important that a new strain is classified into one China lineage based on a nucleotide sequence as soon as possible, and we can further to know the possible epidemic origin and distribution. To be convenient to obtain any nucleotide segment sequence of any China lineage as reference sequence for researchers, we selected some representative rabies strains from each lineages and sequenced their genomes (already published in NCBI GenBank databases). In addition,we can better know the molecular characteristics of different lineages from genome level, and compare the molecular differences of China lineages, even establish molecular basis for further explore the biological characteristics of different lineages.
Until now, total 43 rabies genomes of China street strains are available (Figure 2), including 12 genomes newly sequenced and one Yunnan strain (China VI) reported previously by our lab[12]. Mostly responsible for the current rabies epidemic, China I lineage owns mass of the genomes (27/43), with 93.1%-99.8% genome similarities. China II (8/43)includes 8 genomes, and half of them are ferret badger strains since most of ferret badger rabies belonged to China II clade[16].
China III was often found in wildlife (Raccoon dog, Red fox, et al.) in Inner Mongolia in recent years,threatening human and domestic animals' safety in this region[17-18]. Much distance for year and region between the earlier strains (DRV and MRV) and Inner Mongolia strains may result in lower genome similarities (90%-99.1%).
Although Arctic-like rabies virus (corresponding to China IV) was reported in Inner Mongolia in 2011[17], and then was reported in Qinghai and Tibet in 2015, no relative genomes are available[7]. In this study, we sequenced and reported the genome of Qinghai strain (CQH1202D), not only compared with other lineages, but also compared with all the Arctic and Arctic-like genomes (Figure 3). Based on the N gene phylogenetic analysis previously[7,17], we already knew that China IV lineage had close relationship with strains from South Korea and Russia, equivalent to Arctic-like-2 in global studies of rabies. The genome homology results also consistent,and Qinghai strain had about 95% genome similarities with three South Korea strains[19], only about 91% with 6 South Asia strains (corresponding to Arctic-like-1)[20].
China V is the only lineage not to be detected by our lab in the surveillance program, just was found in Ningxia and Chongqing provinces before 2005[2,15]. Specifically, the genome size of China V (11908nt) is obviously shorter than other lineages' (11923-11925nt) for N-P non-coding regions of China V lineage (73nt) much shorter than other lineages' (90 or 91nt). In the Lyssavirus genus, only a bat species (West Caucasion bat virus, WCBV) had a close N-P size (64nt)[21]. Whether the obvious differences in genomes can influence the biological characteristics even the epidemic, is worth to explore further.
China VI now just was restricted in Yunnan and Guangxi provinces[2], and our lab specially reported the only genome (CYN1009D strain) of China VI previously[12].
In conclusion, we supplemented the rabies genome data of China, and ensured at least one genome was available in each China lineage,moreover, the molecular characteristics of 6 different China lineages were compared and analyzed from genome level, which is a very important part of our rabies surveillance program,and will be much convenient to rapidly fix local rabies emergence or reemergence origin, and benefits for continued comprehensive rabies surveillance, rabies prevention and control in China.
Authors' Contributions LH drafted the manuscript. GZY and ZJ sequenced the 12 genomes. TQ and LHT revised the manuscript in detail. TXY and ZWY provided some epidemic data and backgrounds.
Accepted: April 25, 2016
REFERENCES
1. Hu R, Tang Q, Tang J, et al. Rabies in China: an update. Vector Borne Zoonotic Dis, 2009; 9, 1-12.
2. Tao XY, Tang Q, Rayner S, et al. Molecular Phylodynamic Analysis Indicates Lineage Displacement Occurred in Chinese Rabies Epidemics between 1949 to 2010. PLoS Negl Trop Dis,2013; 7, e2294.
3. Tang X, Luo M, Zhang S, et al. Pivotal role of dogs in rabies transmission, China. Emerg Infect Dis, 2005; 11, 1970-2.
4. Yin CP, Zhou H, Wu H, et al. Analysis on factors related to rabies epidemic in China from 2007-2011. Virol Sin, 2012; 27,132-43.
5. Song M, Tang Q, Rayner S, et al. Human rabies surveillance and control in China, 2005-2012. BMC Infect Dis, 2014; 14, 212.
6. Lang SL, Tao XY, Guo ZY, et al. Molecular characterization of viral G gene in emerging and Re-emerging areas of rabies in China, 2007 to 2011. Virol Sin, 2012; 27, 194-203.
7. Tao XY, Guo ZY, Li H, et al. Rabies Cases in the West of China Have Two Distinct Origins. PLoS Negl Trop Dis, 2015; 9, e0004140.
8. Liu Y, Li N, Zhang S, et al. Analysis of the complete genome of the first Irkut virus isolate from China: Comparison across the Lyssavirus genus. Mol Phylogenet Evol, 2013; 18.
9. Guo Z, Tao X, Yin C, et al. National borders effectively halt the spread of rabies: the current rabies epidemic in china is dislocated from cases in neighboring countries. PLoS Negl Trop Dis, 2013; 7, e2039.
10.Yu J, Li H, Tang Q, et al. The spatial and temporal dynamics of rabies in china. PLoS Negl Trop Dis, 2012; 6, e1640.
11.Tao XY, Tang Q, Li H, et al. Molecular epidemiology of rabies in Southern People's Republic of China. Emerg Infect Dis, 2009;15, 1192-8.
12.Zhang J, Zhang HL, Tao XY, et al. The full-length genome analysis of a street rabies virus strain isolated in Yunnan province of China. Virol Sin, 2012; 27, 204-13.
13.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics, 2007; 23, 2947-8.
14.Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance, and maximum parsimony methods. Mol Biol Evol, 2011; 28, 2731-9.
15.He CQ, Meng SL, Yan HY, et al. Isolation and identification of a novel rabies virus lineage in China with natural recombinant nucleoprotein gene. PLoS One, 2012; 7, e49992.
16.Liu Y, Zhang S, Wu X, et al. Ferret badger rabies origin and its revisited importance as potential source of rabies transmission in Southeast China. BMC Infect Dis, 2010; 10, 234.
17.Shao XQ, Yan XJ, Luo GL, et al. Genetic evidence for domestic raccoon dog rabies caused by Arctic-like rabies virus in Inner Mongolia, China. Epidemiol Infect, 2011; 139, 629-35.
18.Feng Y, Wang W, Guo J, et al. Disease outbreaks caused by steppe-type rabies viruses in China. Epidemiol Infect, 2014; 31,1-5.
19.Oem JK, Kim SH, Kim YH, et al. Complete genome sequences of three rabies viruses isolated from rabid raccoon dogs and a cow in Korea. Virus Genes, 2013; 47, 563-8.
20.Kuzmin IV, Hughes GJ, Botvinkin AD, et al. Arctic and Arctic-like rabies viruses: distribution, phylogeny and evolutionary history.Epidemiol Infect, 2008; 136, 509-19.
21.Tao X, Han N, Guo Z, et al. Molecular characterization of China human rabies vaccine strains. Virol Sin, 2013; 28, 116-23.
Biomed Environ Sci, 2016; 29(5): 340-346 10.3967/bes2016.044 ISSN: 0895-3988
*This work was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (No.2012ZX10004215).
#Correspondence should be addressed to LIU Hong Tu, Tel: 86-10-58900888, E-mail: hotliu@gmail.com; TANG Qing, Tel:86-10-58900895, E-mail: qtang04@sina.com
Biographical note of the first author: LI Hao, male, born in 1981, Associate Researcher, majoring in establishing the laboratory detection system and molecular epidemiology investigation of rabies and arbovirus.
February 21, 2016;
 Biomedical and Environmental Sciences2016年5期
Biomedical and Environmental Sciences2016年5期
- Biomedical and Environmental Sciences的其它文章
- Development of a Novel PmpD-N ELISA for Chlamydia psittaci Infection*
- Evaluation of Six Recombinant Proteins for Serological Diagnosis of Lyme Borreliosis in China*
- Viral Etiology Relationship between Human Papillomavirus and Human Breast Cancer and Target of Gene Therapy
- The Status and Associated Factors of Successful Aging among Older Adults Residing in Longevity Areas in China*
- Cognitive Training in Older Adults with Mild Cognitive Impairment
- Dietary Exposure to Benzyl Butyl Phthalate in China*
